Metagenomic analysis of medicinal Cannabis samples; pathogenic bacteria, toxigenic fungi, and beneficial microbes grow in culture-based yeast and mold tests
- PMID: 27853518
- PMCID: PMC5089129
- DOI: 10.12688/f1000research.9662.1
Metagenomic analysis of medicinal Cannabis samples; pathogenic bacteria, toxigenic fungi, and beneficial microbes grow in culture-based yeast and mold tests
Abstract
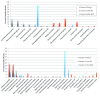
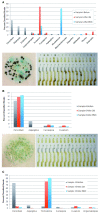
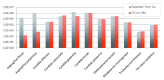
Background: The presence of bacteria and fungi in medicinal or recreational Cannabis poses a potential threat to consumers if those microbes include pathogenic or toxigenic species. This study evaluated two widely used culture-based platforms for total yeast and mold (TYM) testing marketed by 3M Corporation and Biomérieux, in comparison with a quantitative PCR (qPCR) approach marketed by Medicinal Genomics Corporation. Methods: A set of 15 medicinal Cannabis samples were analyzed using 3M and Biomérieux culture-based platforms and by qPCR to quantify microbial DNA. All samples were then subjected to next-generation sequencing and metagenomics analysis to enumerate the bacteria and fungi present before and after growth on culture-based media. Results: Several pathogenic or toxigenic bacterial and fungal species were identified in proportions of >5% of classified reads on the samples, including Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, Ralstonia pickettii, Salmonella enterica, Stenotrophomonas maltophilia, Aspergillus ostianus, Aspergillus sydowii, Penicillium citrinum and Penicillium steckii. Samples subjected to culture showed substantial shifts in the number and diversity of species present, including the failure of Aspergillus species to grow well on either platform. Substantial growth of Clostridium botulinum and other bacteria were frequently observed on one or both of the culture-based TYM platforms. The presence of plant growth promoting (beneficial) fungal species further influenced the differential growth of species in the microbiome of each sample. Conclusions: These findings have important implications for the Cannabis and food safety testing industries.
Keywords: 3M-Petrifilm; Biomérieux-TEMPO; Cannabis; Illumina; PathogINDICAtor-qPCR; metagenomics; microbiome; safety.
Conflict of interest statement
KM, JS, YH, RCL, AD-L, LZ, WO, JW, TF and DRS are employees of Courtagen Life Sciences, the parent company of Medicinal Genomics, which manufactures the commercial qPCR test used in this study.
Figures
References
-
- Compant S, Clément C, Sessitsch A: Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42(5):669–78. 10.1016/j.soilbio.2009.11.024 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

