Chromosomal Amplification of the blaOXA-58 Carbapenemase Gene in a Proteus mirabilis Clinical Isolate
- PMID: 27855079
- PMCID: PMC5278702
- DOI: 10.1128/AAC.01697-16
Chromosomal Amplification of the blaOXA-58 Carbapenemase Gene in a Proteus mirabilis Clinical Isolate
Abstract
Horizontal gene transfer may occur between distantly related bacteria, thus leading to genetic plasticity and in some cases to acquisition of novel resistance traits. Proteus mirabilis is an enterobacterial species responsible for human infections that may express various acquired β-lactam resistance genes, including different classes of carbapenemase genes. Here we report a Proteus mirabilis clinical isolate (strain 1091) displaying resistance to penicillin, including temocillin, together with reduced susceptibility to carbapenems and susceptibility to expanded-spectrum cephalosporins. Using biochemical tests, significant carbapenem hydrolysis was detected in P. mirabilis 1091. Since PCR failed to detect acquired carbapenemase genes commonly found in Enterobacteriaceae, we used a whole-genome sequencing approach that revealed the presence of blaOXA-58 class D carbapenemase gene, so far identified only in Acinetobacter species. This gene was located on a 3.1-kb element coharboring a blaAmpC-like gene. Remarkably, these two genes were bracketed by putative XerC-XerD binding sites and inserted at a XerC-XerD site located between the terminase-like small- and large-subunit genes of a bacteriophage. Increased expression of the two bla genes resulted from a 6-time tandem amplification of the element as revealed by Southern blotting. This is the first isolation of a clinical P. mirabilis strain producing OXA-58, a class D carbapenemase, and the first description of a XerC-XerD-dependent insertion of antibiotic resistance genes within a bacteriophage. This study revealed a new role for the XerC-XerD recombinase in bacteriophage biology.
Keywords: Acinetobacter baumannii; OXA-58; Proteus mirabilis; XerC-XerD recombinase; bacteriophage; carbapenems.
Copyright © 2017 American Society for Microbiology.
Figures
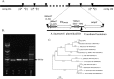
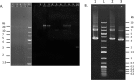

References
-
- Tibbetts R, Frye JG, Marschall J, Warren D, Dunne W. 2008. Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC beta-lactamase in P. mirabilis. J Clin Microbiol 46:3080–3083. doi: 10.1128/JCM.00979-08. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

