Epigenetic regulation of intestinal stem cells by Tet1-mediated DNA hydroxymethylation
- PMID: 27856615
- PMCID: PMC5131782
- DOI: 10.1101/gad.288035.116
Epigenetic regulation of intestinal stem cells by Tet1-mediated DNA hydroxymethylation
Abstract
Methylated cytosines are associated with gene silencing. The ten-eleven translocation (TET) hydroxylases, which oxidize methylated cytosines to 5-hydroxymethylcytosine (5hmC), are essential for cytosine demethylation. Gene silencing and activation are critical for intestinal stem cell (ISC) maintenance and differentiation, but the potential role of TET hydroxylases in these processes has not yet been examined. Here, we generated genome-wide maps of the 5hmC mark in ISCs and their differentiated progeny. Genes with high levels of hydroxymethylation in ISCs are strongly associated with Wnt signaling and developmental processes. We found Tet1 to be the most abundantly expressed Tet gene in ISCs; therefore, we analyzed intestinal development in Tet1-deficient mice and determined that these mice are growth-retarded, exhibit partial postnatal lethality, and have significantly reduced numbers of proliferative cells in the intestinal epithelium. In addition, the Tet1-deficient intestine displays reduced organoid-forming capacity. In the Tet1-deficient crypt, decreased expression of Wnt target genes such as Axin2 and Lgr5 correlates with lower 5hmC levels at their promoters. These data demonstrate that Tet1-mediated DNA hydroxymethylation plays a critical role in the epigenetic regulation of the Wnt pathway in intestinal stem and progenitor cells and consequently in the self-renewal of the intestinal epithelium.
Keywords: Tet1; epigenomics; hydroxymethylation; intestinal differentiation; intestinal stem cell.
© 2016 Kim et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

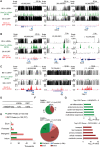

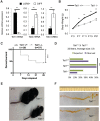

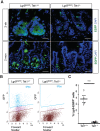
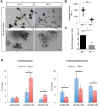
References
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007. - PubMed
-
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 10: 687–692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
