Lineage-specific roles of the cytoplasmic polyadenylation factor CPEB4 in the regulation of melanoma drivers
- PMID: 27857118
- PMCID: PMC5120223
- DOI: 10.1038/ncomms13418
Lineage-specific roles of the cytoplasmic polyadenylation factor CPEB4 in the regulation of melanoma drivers
Abstract
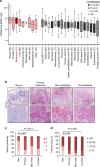
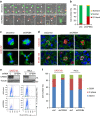
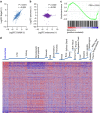
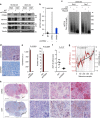
Nuclear 3'-end-polyadenylation is essential for the transport, stability and translation of virtually all eukaryotic mRNAs. Poly(A) tail extension can also occur in the cytoplasm, but the transcripts involved are incompletely understood, particularly in cancer. Here we identify a lineage-specific requirement of the cytoplasmic polyadenylation binding protein 4 (CPEB4) in malignant melanoma. CPEB4 is upregulated early in melanoma progression, as defined by computational and histological analyses. Melanoma cells are distinct from other tumour cell types in their dependency on CPEB4, not only to prevent mitotic aberrations, but to progress through G1/S cell cycle checkpoints. RNA immunoprecipitation, sequencing of bound transcripts and poly(A) length tests link the melanoma-specific functions of CPEB4 to signalling hubs specifically enriched in this disease. Essential in these CPEB4-controlled networks are the melanoma drivers MITF and RAB7A, a feature validated in clinical biopsies. These results provide new mechanistic links between cytoplasmic polyadenylation and lineage specification in melanoma.
Figures





References
-
- Weill L., Belloc E., Bava F. A. & Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 19, 577–585 (2012). - PubMed
-
- Mendez R. & Richter J. D. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2, 521–529 (2001). - PubMed
-
- D'Ambrogio A., Nagaoka K. & Richter J. D. Translational control of cell growth and malignancy by the CPEBs. Nat. Rev. Cancer 13, 283–290 (2013). - PubMed
-
- Fernandez-Miranda G. & Mendez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res. Rev. 11, 460–472 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

