Assessment of Circulating LncRNAs Under Physiologic and Pathologic Conditions in Humans Reveals Potential Limitations as Biomarkers
- PMID: 27857151
- PMCID: PMC5114641
- DOI: 10.1038/srep36596
Assessment of Circulating LncRNAs Under Physiologic and Pathologic Conditions in Humans Reveals Potential Limitations as Biomarkers
Abstract
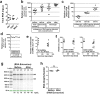
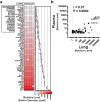
Long non-coding RNAs (lncRNA) are a new class of regulatory molecules with diverse cellular functions. Recent reports have suggested that extracellular lncRNAs are detectable in human plasma and may serve as biomarkers. Here, we sought to investigate circulating lncRNAs as potential biomarkers for pulmonary arterial hypertension (PAH). Eighty-four lncRNAs, representing some of the most abundant and functionally relevant candidates identified in cellular studies, were assessed via RT-qPCR in plasma from PAH and healthy subjects. However, despite preamplification, the majority of lncRNAs were surprisingly undetectable or sporadically detectable, and showed no differential changes. Systematic characterization of plasma/RNA quality and technical performance via internal and external controls revealed no evidence of RNA degradation or RT-qPCR inhibition, and most lncRNAs were robustly detectable in pulmonary tissue. In plasma, lncRNA levels were the lowest among several different RNA species examined, and this was generalizable to other chronic and acute vascular conditions including coronary artery disease, acute coronary syndrome, and septic shock. In addition, two of three previously reported circulating lncRNA biomarker candidates were not detectable in any of the plasma samples. This study reveals new insight on the relative levels of lncRNAs in circulation, which has important implications for their potential development as biomarkers.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

