Mechanism of muscle contraction based on stochastic properties of single actomyosin motors observed in vitro
- PMID: 27857548
- PMCID: PMC5036627
- DOI: 10.2142/biophysics.1.1
Mechanism of muscle contraction based on stochastic properties of single actomyosin motors observed in vitro
Abstract
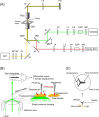
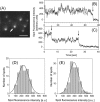
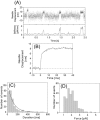
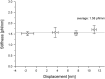
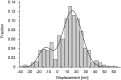
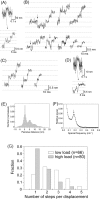
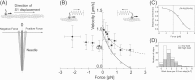
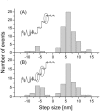
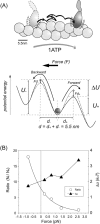
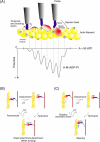
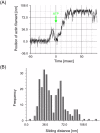
We have previously measured the process of displacement generation by a single head of muscle myosin (S1) using scanning probe nanometry. Given that the myosin head was rigidly attached to a fairly large scanning probe, it was assumed to stably interact with an underlying actin filament without diffusing away as would be the case in muscle. The myosin head has been shown to step back and forth stochastically along an actin filament with actin monomer repeats of 5.5 nm and to produce a net movement in the forward direction. The myosin head underwent 5 forward steps to produce a maximum displacement of 30 nm per ATP at low load (<1 pN). Here, we measured the steps over a wide range of forces up to 4 pN. The size of the steps (∼5.5 nm) did not change as the load increased whereas the number of steps per displacement and the stepping rate both decreased. The rate of the 5.5-nm steps at various force levels produced a force-velocity curve of individual actomyosin motors. The force-velocity curve from the individual myosin heads was comparable to that reported in muscle, suggesting that the fundamental mechanical properties in muscle are basically due to the intrinsic stochastic nature of individual actomyosin motors. In order to explain multiple stochastic steps, we propose a model arguing that the thermally-driven step of a myosin head is biased in the forward direction by a potential slope along the actin helical pitch resulting from steric compatibility between the binding sites of actin and a myosin head. Furthermore, computer simulations show that multiple cooperating heads undergoing stochastic steps generate a long (>60 nm) sliding distance per ATP between actin and myosin filaments, i.e., the movement is loosely coupled to the ATPase cycle as observed in muscle.
Keywords: Brownian motion; loose coupling; molecular motor; preferential landing; single molecule.
Figures

Similar articles
-
Coupling of lever arm swing and biased Brownian motion in actomyosin.PLoS Comput Biol. 2014 Apr 24;10(4):e1003552. doi: 10.1371/journal.pcbi.1003552. eCollection 2014 Apr. PLoS Comput Biol. 2014. PMID: 24762409 Free PMC article.
-
Cooperative actions between myosin heads bring effective functions.Biosystems. 2007 Apr;88(3):293-300. doi: 10.1016/j.biosystems.2006.03.013. Epub 2006 Nov 10. Biosystems. 2007. PMID: 17187925
-
Multiple- and single-molecule analysis of the actomyosin motor by nanometer-piconewton manipulation with a microneedle: unitary steps and forces.Biophys J. 1996 Jan;70(1):383-400. doi: 10.1016/S0006-3495(96)79582-6. Biophys J. 1996. PMID: 8770215 Free PMC article.
-
Loose coupling between chemical and mechanical reactions in actomyosin energy transduction.Adv Biophys. 1990;26:75-95. doi: 10.1016/0065-227x(90)90008-h. Adv Biophys. 1990. PMID: 2082730 Review.
-
Induced potential model of muscular contraction mechanism and myosin molecular structure.Adv Biophys. 1999;36:107-58. doi: 10.1016/s0065-227x(99)80006-9. Adv Biophys. 1999. PMID: 10463074 Review.
Cited by
-
Landscape-Based View on the Stepping Movement of Myosin VI.J Phys Chem B. 2022 Sep 29;126(38):7262-7270. doi: 10.1021/acs.jpcb.2c03694. Epub 2022 Sep 15. J Phys Chem B. 2022. PMID: 36107864 Free PMC article.
-
Coupling of lever arm swing and biased Brownian motion in actomyosin.PLoS Comput Biol. 2014 Apr 24;10(4):e1003552. doi: 10.1371/journal.pcbi.1003552. eCollection 2014 Apr. PLoS Comput Biol. 2014. PMID: 24762409 Free PMC article.
-
Single molecule detection, thermal fluctuation and life.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(2):51-63. doi: 10.2183/pjab.93.004. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28190869 Free PMC article. Review.
-
Distinct Conformation of ATP Molecule in Solution and on Protein.Biophysics (Nagoya-shi). 2013 Jan 18;9:1-12. doi: 10.2142/biophysics.9.1. eCollection 2013. Biophysics (Nagoya-shi). 2013. PMID: 27493535 Free PMC article.
-
From single molecule fluctuations to muscle contraction: a Brownian model of A.F. Huxley's hypotheses.PLoS One. 2012;7(7):e40042. doi: 10.1371/journal.pone.0040042. Epub 2012 Jul 16. PLoS One. 2012. PMID: 22815722 Free PMC article.
References
-
- Huxley HE, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. - PubMed
-
- Huxley AF, Niedergerke R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature. 1954;173:971–973. - PubMed
-
- Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. - PubMed
-
- Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. - PubMed
-
- Yanagida T. Angles of nucleotides bound to cross-bridges in glycerinated muscle fiber at various concentrations ε-ATP, ε-ADP and ε-AMPPNP detected by polarized fluorescence. J. Mol. Biol. 1981;146:539–560. - PubMed
LinkOut - more resources
Full Text Sources
