Ratio of membrane proteins in total proteomes of prokaryota
- PMID: 27857565
- PMCID: PMC5036657
- DOI: 10.2142/biophysics.3.37
Ratio of membrane proteins in total proteomes of prokaryota
Abstract
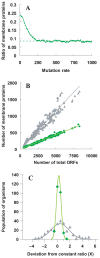
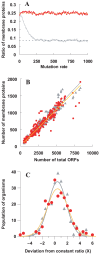
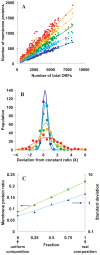
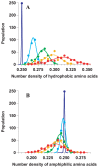
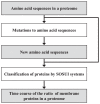
The numbers of membrane proteins in the current genomes of various organisms provide an important clue about how the protein world has evolved from the aspect of membrane proteins. Numbers of membrane proteins were estimated by analyzing the total proteomes of 248 prokaryota, using the SOSUI system for membrane proteins (Hirokawa et al., Bioinformatics, 1998) and SOSUI-signal for signal peptides (Gomi et al., CBIJ, 2004). The results showed that the ratio of membrane proteins to total proteins in these proteomes was almost constant: 0.228. When amino acid sequences were randomized, setting the probability of occurrence of all amino acids to 5%, the membrane protein/total protein ratio decreased to about 0.085. However, when the same simulation was carried out, but using the amino acid composition of the above proteomes, this ratio was 0.218, which is nearly the same as that of the real proteomic systems. This fact is consistent with the birth, death and innovation (BDI) model for membrane proteins, in which transmembrane segments emerge and disappear in accordance with random mutation events.
Keywords: comparative proteomics; large-scale genome comparison; membrane protein prediction; protein world; sequence simulation.
Figures


References
-
- Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–1703. - PubMed
-
- Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. - PubMed
-
- Qian J, Luscombe NM, Gerstein M. Protein family and fold occurrence in genomes: power-law behaviour and evolutionary model. J Mol Biol. 2001;313:673–681. - PubMed
-
- Huynen MA, van Nimwegen E. The frequency distribution of gene family sizes in complete genomes. Mol Biol Evol. 1998;15:583–589. - PubMed
-
- Vogel C, Teichmann SA, Pereira-Leal J. The relationship between domain duplication and recombination. J Mol Biol. 2005;346:355–365. - PubMed
LinkOut - more resources
Full Text Sources
