Insight into the sequence specificity of a probe on an Affymetrix GeneChip by titration experiments using only one oligonucleotide
- PMID: 27857566
- PMCID: PMC5036658
- DOI: 10.2142/biophysics.3.47
Insight into the sequence specificity of a probe on an Affymetrix GeneChip by titration experiments using only one oligonucleotide
Abstract
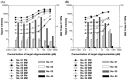
High-density oligonucleotide arrays are powerful tools for the analysis of genome-wide expression of genes and for genome-wide screens of genetic variation in living organisms. One of the critical problems in high-density oligonucleotide arrays is how to identify the actual amounts of a transcript due to noise and cross-hybridization involved in the observed signal intensities. Although mismatch (MM) probes are spotted on Affymetrix GeneChips to evaluate the noise and cross-hybridization embedded in perfect match (PM) probes, the behavior of probe-level signal intensities remains unclear. In the present study, we hybridized only one complement 25-mer oligonucleotide to characterize the behavior of duplex formation between target and probe in the complete absence of cross-hybridization. Titration experiments using only one oligonucleotide demonstrated that a substantial amount of intact target was hybridized not only to the PM but also the MM probe and that duplex formation between intact target and MM probe was efficiently reduced by increasing the stringency of hybridization conditions and shortening probe length. In addition, we discuss the correlation between potential for secondary structure of target oligonucleotide and hybridization intensity. These findings will be useful for the development of genome-wide analysis of gene expression and genetic variations by optimization of hybridization and probe conditions.
Keywords: cross-hybridization; hybridization; microarray; probe length; probe-level signal.
Figures



Similar articles
-
Experimental optimization of probe length to increase the sequence specificity of high-density oligonucleotide microarrays.BMC Genomics. 2007 Oct 16;8:373. doi: 10.1186/1471-2164-8-373. BMC Genomics. 2007. PMID: 17939865 Free PMC article.
-
A simple optimization can improve the performance of single feature polymorphism detection by Affymetrix expression arrays.BMC Genomics. 2010 May 20;11:315. doi: 10.1186/1471-2164-11-315. BMC Genomics. 2010. PMID: 20482895 Free PMC article.
-
Empirical establishment of oligonucleotide probe design criteria.Appl Environ Microbiol. 2005 Jul;71(7):3753-60. doi: 10.1128/AEM.71.7.3753-3760.2005. Appl Environ Microbiol. 2005. PMID: 16000786 Free PMC article.
-
Affymetrix oligonucleotide analysis of gene expression in the injured heart.Methods Mol Med. 2005;112:305-20. doi: 10.1385/1-59259-879-x:305. Methods Mol Med. 2005. PMID: 16010026 Review.
-
Using oligonucleotide probe arrays to access genetic diversity.Biotechniques. 1995 Sep;19(3):442-7. Biotechniques. 1995. PMID: 7495558 Review.
Cited by
-
A comparison of the TempO-Seq and Affymetrix microarray platform using RTqPCR validation.BMC Genomics. 2024 Jul 3;25(1):669. doi: 10.1186/s12864-024-10586-7. BMC Genomics. 2024. PMID: 38961363 Free PMC article.
-
An improved physico-chemical model of hybridization on high-density oligonucleotide microarrays.Bioinformatics. 2008 May 15;24(10):1278-85. doi: 10.1093/bioinformatics/btn109. Epub 2008 Mar 31. Bioinformatics. 2008. PMID: 18378525 Free PMC article.
References
-
- Noordewier MO, Warren PV. Gene expression microarrays and the integration of biological knowledge. Trends Biotechnol. 2001;19:412–415. - PubMed
-
- Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21:42–47. - PubMed
-
- Mir KU, Southern EM. Sequence variation in genes and genomic DNA: methods for large-scale analysis. Annu Rev Genomics Hum Genet. 2000;1:329–360. - PubMed
-
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. - PubMed
-
- Affymetrix Microarray Suite 5.0 User’s Guide. 2001.
LinkOut - more resources
Full Text Sources
Miscellaneous
