Profile conditional random fields for modeling protein families with structural information
- PMID: 27857577
- PMCID: PMC5036637
- DOI: 10.2142/biophysics.5.37
Profile conditional random fields for modeling protein families with structural information
Abstract
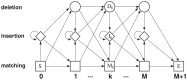
A statistical model of protein families, called profile conditional random fields (CRFs), is proposed. This model may be regarded as an integration of the profile hidden Markov model (HMM) and the Finkelstein-Reva (FR) theory of protein folding. While the model structure of the profile CRF is almost identical to the profile HMM, it can incorporate arbitrary correlations in the sequences to be aligned to the model. In addition, like in the FR theory, the profile CRF can incorporate long-range pair-wise interactions between model states via mean-field-like approximations. We give the detailed formulation of the model, self-consistent approximations for treating long-range interactions, and algorithms for computing partition functions and marginal probabilities. We also outline the methods for the global optimization of model parameters as well as a Bayesian framework for parameter learning and selection of optimal alignments.
Keywords: dynamic programming; fold recognition; mean field approximation; sequence analysis; structure prediction.
Figures
Similar articles
-
HMM-ModE--improved classification using profile hidden Markov models by optimising the discrimination threshold and modifying emission probabilities with negative training sequences.BMC Bioinformatics. 2007 Mar 27;8:104. doi: 10.1186/1471-2105-8-104. BMC Bioinformatics. 2007. PMID: 17389042 Free PMC article.
-
The infinite-order conditional random field model for sequential data modeling.IEEE Trans Pattern Anal Mach Intell. 2013 Jun;35(6):1523-34. doi: 10.1109/TPAMI.2012.208. IEEE Trans Pattern Anal Mach Intell. 2013. PMID: 23599063
-
Protein fold recognition using segmentation conditional random fields (SCRFs).J Comput Biol. 2006 Mar;13(2):394-406. doi: 10.1089/cmb.2006.13.394. J Comput Biol. 2006. PMID: 16597248
-
ProbPFP: a multiple sequence alignment algorithm combining hidden Markov model optimized by particle swarm optimization with partition function.BMC Bioinformatics. 2019 Nov 25;20(Suppl 18):573. doi: 10.1186/s12859-019-3132-7. BMC Bioinformatics. 2019. PMID: 31760933 Free PMC article.
-
Hidden Markov models.Curr Opin Struct Biol. 1996 Jun;6(3):361-5. doi: 10.1016/s0959-440x(96)80056-x. Curr Opin Struct Biol. 1996. PMID: 8804822 Review.
Cited by
-
A unified statistical model of protein multiple sequence alignment integrating direct coupling and insertions.Biophys Physicobiol. 2016 Apr 22;13:45-62. doi: 10.2142/biophysico.13.0_45. eCollection 2016. Biophys Physicobiol. 2016. PMID: 27924257 Free PMC article.
References
-
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. - PubMed
-
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. - PubMed
-
- Gotoh O. An improved algorithm for matching biological sequences. J Mol Biol. 1982;162:705–708. - PubMed
-
- Durbin R, Eddy R, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge Univ. Press; Cambridge, U.K: 1999.
-
- Eidhammer I, Jonassen I, Taylor WR. Protein bioinformatics. Wiley & Sons; Chichester, England: 2004.
LinkOut - more resources
Full Text Sources