Structure of centromere chromatin: from nucleosome to chromosomal architecture
- PMID: 27858158
- PMCID: PMC5509776
- DOI: 10.1007/s00412-016-0620-7
Structure of centromere chromatin: from nucleosome to chromosomal architecture
Abstract
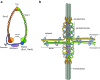
The centromere is essential for the segregation of chromosomes, as it serves as attachment site for microtubules to mediate chromosome segregation during mitosis and meiosis. In most organisms, the centromere is restricted to one chromosomal region that appears as primary constriction on the condensed chromosome and is partitioned into two chromatin domains: The centromere core is characterized by the centromere-specific histone H3 variant CENP-A (also called cenH3) and is required for specifying the centromere and for building the kinetochore complex during mitosis. This core region is generally flanked by pericentric heterochromatin, characterized by nucleosomes containing H3 methylated on lysine 9 (H3K9me) that are bound by heterochromatin proteins. During mitosis, these two domains together form a three-dimensional structure that exposes CENP-A-containing chromatin to the surface for interaction with the kinetochore and microtubules. At the same time, this structure supports the tension generated during the segregation of sister chromatids to opposite poles. In this review, we discuss recent insight into the characteristics of the centromere, from the specialized chromatin structures at the centromere core and the pericentromere to the three-dimensional organization of these regions that make up the functional centromere.
Keywords: CENP-A; Centromere; Chromatin; Cohesin; Pericentromere.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

