Local adaptation at higher trophic levels: contrasting hyperparasite-pathogen infection dynamics in the field and laboratory
- PMID: 27859910
- PMCID: PMC5412677
- DOI: 10.1111/mec.13928
Local adaptation at higher trophic levels: contrasting hyperparasite-pathogen infection dynamics in the field and laboratory
Abstract
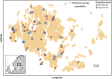
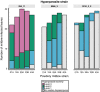
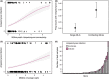
Predicting and controlling infectious disease epidemics is a major challenge facing the management of agriculture, human and wildlife health. Co-evolutionarily derived patterns of local adaptation among pathogen populations have the potential to generate variation in disease epidemiology; however, studies of local adaptation in disease systems have mostly focused on interactions between competing pathogens or pathogens and their hosts. In nature, parasites and pathogens are also subject to attack by hyperparasitic natural enemies that can severely impact upon their infection dynamics. However, few studies have investigated whether this interaction varies across combinations of pathogen-hyperparasite strains, and whether this influences hyperparasite incidence in natural pathogen populations. Here, we test whether the association between a hyperparasitic fungus, Ampelomyces, and a single powdery mildew host, Podosphaera plantaginis, varies among genotype combinations, and whether this drives hyperparasite incidence in nature. Laboratory inoculation studies reveal that genotype, genotype × genotype interactions and local adaptation affect hyperparasite infection. However, observations of a natural pathogen metapopulation reveal that spatial rather than genetic factors predict the risk of hyperparasite presence. Our results highlight how sensitive the outcome of biocontrol using hyperparasites is to selection of hyperparasite strains.
Keywords: co-evolution; disease; host-parasite interactions; hyperparasite; local adaptation.
© 2016 The Authors Molecular Ecology Published by John Wiley & Sons Ltd.
Figures


References
-
- Abo‐Foul S, Raskin VI, Sztejnberg A, Marder JB (1996) Disruption of chlorophyll organization and function in powdery mildew‐diseased cucumber leaves and its control by the hyperparasite Ampelomyces quisqualis . Phytopathology, 86, 195–199.
-
- Anderson RM, May RM (1991) Infectious Diseases of Humans: Dynamics and Control. Oxford University Press, Oxford.
-
- Angeli D, Maurhofer M, Gessler C, Pertot I (2012a) Existence of different physiological forms within genetically diverse strains of Ampelomyces quisqualis . Phytoparasitica, 40, 37–51.
-
- Angeli D, Puopolo G, Maurhofer M, Gessler C, Pertot I (2012b) Is the mycoparasitic activity of Ampelomyces quisqualis biocontrol strains related to phylogeny and hydrolytic enzyme production? Biological Control, 63, 348–358.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

