Two Opsin 3-Related Proteins in the Chicken Retina and Brain: A TMT-Type Opsin 3 Is a Blue-Light Sensor in Retinal Horizontal Cells, Hypothalamus, and Cerebellum
- PMID: 27861495
- PMCID: PMC5115664
- DOI: 10.1371/journal.pone.0163925
Two Opsin 3-Related Proteins in the Chicken Retina and Brain: A TMT-Type Opsin 3 Is a Blue-Light Sensor in Retinal Horizontal Cells, Hypothalamus, and Cerebellum
Abstract
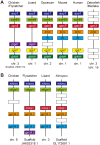
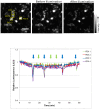
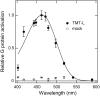
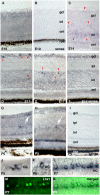
Opsin family genes encode G protein-coupled seven-transmembrane proteins that bind a retinaldehyde chromophore in photoreception. Here, we sought potential as yet undescribed avian retinal photoreceptors, focusing on Opsin 3 homologs in the chicken. We found two Opsin 3-related genes in the chicken genome: one corresponding to encephalopsin/panopsin (Opn3) in mammals, and the other belonging to the teleost multiple tissue opsin (TMT) 2 group. Bioluminescence imaging and G protein activation assays demonstrated that the chicken TMT opsin (cTMT) functions as a blue light sensor when forced-expressed in mammalian cultured cells. We did not detect evidence of light sensitivity for the chicken Opn3 (cOpn3). In situ hybridization demonstrated expression of cTMT in subsets of differentiating cells in the inner retina and, as development progressed, predominant localization to retinal horizontal cells (HCs). Immunohistochemistry (IHC) revealed cTMT in HCs as well as in small numbers of cells in the ganglion and inner nuclear layers of the post-hatch chicken retina. In contrast, cOpn3-IR cells were found in distinct subsets of cells in the inner nuclear layer. cTMT-IR cells were also found in subsets of cells in the hypothalamus. Finally, we found differential distribution of cOpn3 and cTMT proteins in specific cells of the cerebellum. The present results suggest that a novel TMT-type opsin 3 may function as a photoreceptor in the chicken retina and brain.
Conflict of interest statement
Olympus Corporation: this commercial affiliation does not alter our adherence to all PLOS ONE policies on sharing data and materials. Pfizer Japan Inc.: this commercial funder does not alter our adherence to PLOS ONE policies on sharing data and materials, either.
Figures






Similar articles
-
Expression patterns of the opsin 5-related genes in the developing chicken retina.Dev Dyn. 2008 Jul;237(7):1910-22. doi: 10.1002/dvdy.21611. Dev Dyn. 2008. PMID: 18570255
-
Molecular cloning, localization and circadian expression of chicken melanopsin (Opn4): differential regulation of expression in pineal and retinal cell types.J Neurochem. 2005 Jan;92(1):158-70. doi: 10.1111/j.1471-4159.2004.02874.x. J Neurochem. 2005. PMID: 15606905
-
Differential regulation of melatonin synthesis genes and phototransduction genes in embryonic chicken retina and cultured retinal precursor cells.Mol Vis. 2005 Jul 7;11:472-81. Mol Vis. 2005. PMID: 16030498
-
The opsins.Genome Biol. 2005;6(3):213. doi: 10.1186/gb-2005-6-3-213. Epub 2005 Mar 1. Genome Biol. 2005. PMID: 15774036 Free PMC article. Review.
-
Non-visual Opsins and Novel Photo-Detectors in the Vertebrate Inner Retina Mediate Light Responses Within the Blue Spectrum Region.Cell Mol Neurobiol. 2022 Jan;42(1):59-83. doi: 10.1007/s10571-020-00997-x. Epub 2020 Nov 24. Cell Mol Neurobiol. 2022. PMID: 33231827 Free PMC article. Review.
Cited by
-
Expression of Non-visual Opsins Opn3 and Opn5 in the Developing Inner Retinal Cells of Birds. Light-Responses in Müller Glial Cells.Front Cell Neurosci. 2019 Aug 16;13:376. doi: 10.3389/fncel.2019.00376. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31474836 Free PMC article.
-
Activation of an Endogenous Opsin 3 Light Receptor Mediates Photo-Relaxation of Pre-Contracting Late Gestation Human Uterine Smooth Muscle Ex Vivo.Reprod Sci. 2020 Sep;27(9):1791-1801. doi: 10.1007/s43032-020-00180-z. Epub 2020 Mar 12. Reprod Sci. 2020. PMID: 32166706 Free PMC article.
-
Airway smooth muscle photorelaxation via opsin receptor activation.Am J Physiol Lung Cell Mol Physiol. 2019 Jan 1;316(1):L82-L93. doi: 10.1152/ajplung.00135.2018. Epub 2018 Oct 4. Am J Physiol Lung Cell Mol Physiol. 2019. PMID: 30284927 Free PMC article.
-
The Gluopsins: Opsins without the Retinal Binding Lysine.Cells. 2022 Aug 6;11(15):2441. doi: 10.3390/cells11152441. Cells. 2022. PMID: 35954284 Free PMC article.
-
The Sensory Shark: High-quality Morphological, Genomic and Transcriptomic Data for the Small-spotted Catshark Scyliorhinus Canicula Reveal the Molecular Bases of Sensory Organ Evolution in Jawed Vertebrates.Mol Biol Evol. 2024 Dec 6;41(12):msae246. doi: 10.1093/molbev/msae246. Mol Biol Evol. 2024. PMID: 39657112 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

