Pathoadaptive Mutations of Escherichia coli K1 in Experimental Neonatal Systemic Infection
- PMID: 27861552
- PMCID: PMC5115809
- DOI: 10.1371/journal.pone.0166793
Pathoadaptive Mutations of Escherichia coli K1 in Experimental Neonatal Systemic Infection
Abstract
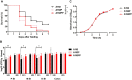
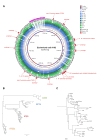
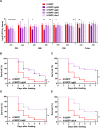
Although Escherichia coli K1 strains are benign commensals in adults, their acquisition at birth by the newborn may result in life-threatening systemic infections, most commonly sepsis and meningitis. Key features of these infections, including stable gastrointestinal (GI) colonization and age-dependent invasion of the bloodstream, can be replicated in the neonatal rat. We previously increased the capacity of a septicemia isolate of E. coli K1 to elicit systemic infection following colonization of the small intestine by serial passage through two-day-old (P2) rat pups. The passaged strain, A192PP (belonging to sequence type 95), induces lethal infection in all pups fed 2-6 x 106 CFU. Here we use whole-genome sequencing to identify mutations responsible for the threefold increase in lethality between the initial clinical isolate and the passaged derivative. Only four single nucleotide polymorphisms (SNPs), in genes (gloB, yjgV, tdcE) or promoters (thrA) involved in metabolic functions, were found: no changes were detected in genes encoding virulence determinants associated with the invasive potential of E. coli K1. The passaged strain differed in carbon source utilization in comparison to the clinical isolate, most notably its inability to metabolize glucose for growth. Deletion of each of the four genes from the E. coli A192PP chromosome altered the proteome, reduced the number of colonizing bacteria in the small intestine and increased the number of P2 survivors. This work indicates that changes in metabolic potential lead to increased colonization of the neonatal GI tract, increasing the potential for translocation across the GI epithelium into the systemic circulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Loss of Trefoil Factor 2 Sensitizes Rat Pups to Systemic Infection with the Neonatal Pathogen Escherichia coli K1.Infect Immun. 2019 Apr 23;87(5):e00878-18. doi: 10.1128/IAI.00878-18. Print 2019 Mar. Infect Immun. 2019. PMID: 30833331 Free PMC article.
-
Genome-Wide Identification by Transposon Insertion Sequencing of Escherichia coli K1 Genes Essential for In Vitro Growth, Gastrointestinal Colonizing Capacity, and Survival in Serum.J Bacteriol. 2018 Mar 12;200(7):e00698-17. doi: 10.1128/JB.00698-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29339415 Free PMC article.
-
Bioluminescent imaging reveals novel patterns of colonization and invasion in systemic Escherichia coli K1 experimental infection in the neonatal rat.Infect Immun. 2015 Dec;83(12):4528-40. doi: 10.1128/IAI.00953-15. Epub 2015 Sep 8. Infect Immun. 2015. PMID: 26351276 Free PMC article.
-
Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2:K1:H5; ST complex 95, ST140) shares close relationship with ST95 APEC O1:K1 and human ExPEC O18:K1 strains.PLoS One. 2014 Nov 14;9(11):e112048. doi: 10.1371/journal.pone.0112048. eCollection 2014. PLoS One. 2014. PMID: 25397580 Free PMC article.
-
Non-invasive three-dimensional imaging of Escherichia coli K1 infection using diffuse light imaging tomography combined with micro-computed tomography.Methods. 2017 Aug 15;127:62-68. doi: 10.1016/j.ymeth.2017.05.005. Epub 2017 May 15. Methods. 2017. PMID: 28522324 Review.
Cited by
-
Loss of Trefoil Factor 2 Sensitizes Rat Pups to Systemic Infection with the Neonatal Pathogen Escherichia coli K1.Infect Immun. 2019 Apr 23;87(5):e00878-18. doi: 10.1128/IAI.00878-18. Print 2019 Mar. Infect Immun. 2019. PMID: 30833331 Free PMC article.
-
Inborn Errors of Immunity in the Premature Infant: Challenges in Recognition and Diagnosis.Front Immunol. 2021 Dec 24;12:758373. doi: 10.3389/fimmu.2021.758373. eCollection 2021. Front Immunol. 2021. PMID: 35003071 Free PMC article. Review.
-
Genome-Wide Identification by Transposon Insertion Sequencing of Escherichia coli K1 Genes Essential for In Vitro Growth, Gastrointestinal Colonizing Capacity, and Survival in Serum.J Bacteriol. 2018 Mar 12;200(7):e00698-17. doi: 10.1128/JB.00698-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29339415 Free PMC article.
-
Phage Targeting Neonatal Meningitis E. coli K1 In Vitro in the Intestinal Microbiota of Pregnant Donors and Impact on Bacterial Populations.Int J Mol Sci. 2023 Jun 24;24(13):10580. doi: 10.3390/ijms241310580. Int J Mol Sci. 2023. PMID: 37445758 Free PMC article.
-
Impact of Sequential Passaging on Protein Expression of E. coli Using Proteomics Analysis.Int J Microbiol. 2020 Jul 30;2020:2716202. doi: 10.1155/2020/2716202. eCollection 2020. Int J Microbiol. 2020. PMID: 32802068 Free PMC article.
References
-
- Robbins JB, McCracken GH, Gotschlich EC, Ørskov F, Ørskov I, Hanson LA. Escherichia coli K1 capsular polysaccharide associated with neonatal meningitis. N Eng J Med. 1974; 290:1216–1220. - PubMed
-
- Kasper DL, Winkelhake JL, Zollinger WD, Brandt BL, Artenstein MS. Immunochemical similarity between polysaccharide antigens of Escherichia coli 07: K1(L):NM and group B Neisseria meningitidis. J Immunol. 1973; 110:262–268. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

