Computational Identification of Tissue-Specific Splicing Regulatory Elements in Human Genes from RNA-Seq Data
- PMID: 27861625
- PMCID: PMC5115852
- DOI: 10.1371/journal.pone.0166978
Computational Identification of Tissue-Specific Splicing Regulatory Elements in Human Genes from RNA-Seq Data
Abstract
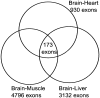
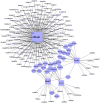
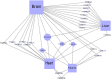
Alternative splicing is a vital process for regulating gene expression and promoting proteomic diversity. It plays a key role in tissue-specific expressed genes. This specificity is mainly regulated by splicing factors that bind to specific sequences called splicing regulatory elements (SREs). Here, we report a genome-wide analysis to study alternative splicing on multiple tissues, including brain, heart, liver, and muscle. We propose a pipeline to identify differential exons across tissues and hence tissue-specific SREs. In our pipeline, we utilize the DEXSeq package along with our previously reported algorithms. Utilizing the publicly available RNA-Seq data set from the Human BodyMap project, we identified 28,100 differentially used exons across the four tissues. We identified tissue-specific exonic splicing enhancers that overlap with various previously published experimental and computational databases. A complicated exonic enhancer regulatory network was revealed, where multiple exonic enhancers were found across multiple tissues while some were found only in specific tissues. Putative combinatorial exonic enhancers and silencers were discovered as well, which may be responsible for exon inclusion or exclusion across tissues. Some of the exonic enhancers are found to be co-occurring with multiple exonic silencers and vice versa, which demonstrates a complicated relationship between tissue-specific exonic enhancers and silencers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Identifying splicing regulatory elements with de Bruijn graphs.J Comput Biol. 2014 Dec;21(12):880-97. doi: 10.1089/cmb.2014.0183. J Comput Biol. 2014. PMID: 25393830 Free PMC article.
-
Computational identification of tissue-specific alternative splicing elements in mouse genes from RNA-Seq.Nucleic Acids Res. 2010 Dec;38(22):7895-907. doi: 10.1093/nar/gkq679. Epub 2010 Aug 4. Nucleic Acids Res. 2010. PMID: 20685814 Free PMC article.
-
Predictive identification of exonic splicing enhancers in human genes.Science. 2002 Aug 9;297(5583):1007-13. doi: 10.1126/science.1073774. Epub 2002 Jul 11. Science. 2002. PMID: 12114529
-
Rules and tools to predict the splicing effects of exonic and intronic mutations.Wiley Interdiscip Rev RNA. 2018 Jan;9(1). doi: 10.1002/wrna.1451. Epub 2017 Sep 26. Wiley Interdiscip Rev RNA. 2018. PMID: 28949076 Review.
-
Splicing mutations in human genetic disorders: examples, detection, and confirmation.J Appl Genet. 2018 Aug;59(3):253-268. doi: 10.1007/s13353-018-0444-7. Epub 2018 Apr 21. J Appl Genet. 2018. PMID: 29680930 Free PMC article. Review.
Cited by
-
mRNA Transcript Variants Expressed in Mammalian Cells.Int J Mol Sci. 2025 Jan 26;26(3):1052. doi: 10.3390/ijms26031052. Int J Mol Sci. 2025. PMID: 39940824 Free PMC article. Review.
-
Polymorphic differences within HLA-C alleles contribute to alternatively spliced transcripts lacking exon 5.HLA. 2022 Sep;100(3):232-243. doi: 10.1111/tan.14695. Epub 2022 Jun 27. HLA. 2022. PMID: 35650170 Free PMC article.
-
The LL-100 panel: 100 cell lines for blood cancer studies.Sci Rep. 2019 Jun 3;9(1):8218. doi: 10.1038/s41598-019-44491-x. Sci Rep. 2019. PMID: 31160637 Free PMC article.
-
The determinants of alternative RNA splicing in human cells.Mol Genet Genomics. 2017 Dec;292(6):1175-1195. doi: 10.1007/s00438-017-1350-0. Epub 2017 Jul 13. Mol Genet Genomics. 2017. PMID: 28707092 Review.
-
Alternative splicing of modulatory immune receptors in T lymphocytes: a newly identified and targetable mechanism for anticancer immunotherapy.Front Immunol. 2025 Jan 7;15:1490035. doi: 10.3389/fimmu.2024.1490035. eCollection 2024. Front Immunol. 2025. PMID: 39845971 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

