Extension of research data repository system to support direct compute access to biomedical datasets: enhancing Dataverse to support large datasets
- PMID: 27862010
- PMCID: PMC5546227
- DOI: 10.1111/nyas.13272
Extension of research data repository system to support direct compute access to biomedical datasets: enhancing Dataverse to support large datasets
Abstract
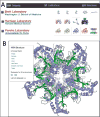
Access to experimental X-ray diffraction image data is important for validation and reproduction of macromolecular models and indispensable for the development of structural biology processing methods. In response to the evolving needs of the structural biology community, we recently established a diffraction data publication system, the Structural Biology Data Grid (SBDG, data.sbgrid.org), to preserve primary experimental datasets supporting scientific publications. All datasets published through the SBDG are freely available to the research community under a public domain dedication license, with metadata compliant with the DataCite Schema (schema.datacite.org). A proof-of-concept study demonstrated community interest and utility. Publication of large datasets is a challenge shared by several fields, and the SBDG has begun collaborating with the Institute for Quantitative Social Science at Harvard University to extend the Dataverse (dataverse.org) open-source data repository system to structural biology datasets. Several extensions are necessary to support the size and metadata requirements for structural biology datasets. In this paper, we describe one such extension-functionality supporting preservation of file system structure within Dataverse-which is essential for both in-place computation and supporting non-HTTP data transfers.
Keywords: Data Access Alliance; Dataverse; RDMS; SBGrid; X-ray diffraction; research data management system.
© 2016 New York Academy of Sciences.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Data publication with the structural biology data grid supports live analysis.Nat Commun. 2016 Mar 7;7:10882. doi: 10.1038/ncomms10882. Nat Commun. 2016. PMID: 26947396 Free PMC article.
-
Big Data access and infrastructure for modern biology: case studies in data repository utility.Ann N Y Acad Sci. 2017 Jan;1387(1):112-123. doi: 10.1111/nyas.13281. Epub 2016 Nov 1. Ann N Y Acad Sci. 2017. PMID: 27801987 Review.
-
Distributed data networks: a blueprint for Big Data sharing and healthcare analytics.Ann N Y Acad Sci. 2017 Jan;1387(1):105-111. doi: 10.1111/nyas.13287. Epub 2016 Nov 18. Ann N Y Acad Sci. 2017. PMID: 27862002 Review.
-
PGP repository: a plant phenomics and genomics data publication infrastructure.Database (Oxford). 2016 Apr 17;2016:baw033. doi: 10.1093/database/baw033. Print 2016. Database (Oxford). 2016. PMID: 27087305 Free PMC article.
-
Data science, learning, and applications to biomedical and health sciences.Ann N Y Acad Sci. 2017 Jan;1387(1):5-11. doi: 10.1111/nyas.13309. Ann N Y Acad Sci. 2017. PMID: 28122121
References
-
- Berman H, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Biol. 2003;10:980–980. - PubMed
-
- Bilderback DH, Elleaume P, Weckert E. Review of third and next generation synchrotron light sources. J Phys B At Mol Opt Phys. 2005;38:S773–S797.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

