Unwinding the twister ribozyme: from structure to mechanism
- PMID: 27863022
- PMCID: PMC5408937
- DOI: 10.1002/wrna.1402
Unwinding the twister ribozyme: from structure to mechanism
Abstract
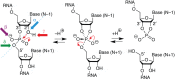
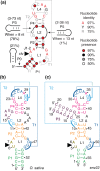
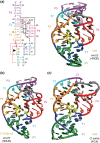
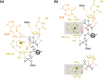
The twister ribozyme motif has been identified by bioinformatic means very recently. Currently, four crystal structures with ordered active sites together with a series of chemical and biochemical data provide insights into how this RNA accomplishes its efficient self-cleavage. Of particular interest for a mechanistic proposal are structural distinctions observed in the active sites that concern the conformation of the U-A cleavage site dinucleotide (in-line alignment of the attacking 2'-O nucleophile to the to-be-cleaved PO5' bond versus suboptimal alignments) as well as the presence/absence of Mg2+ ions at the scissile phosphate. All structures support the notion that an active site guanine and the conserved adenine at the cleavage site are important contributors to cleavage chemistry, likely being involved in general acid base catalysis. Evidence for innersphere coordination of a Mg2+ ion to the pro-S nonbridging oxygen of the scissile phosphate stems from two of the four crystal structures. Together with the finding of thio/rescue effects for phosphorothioate substrates, this suggests the participation of divalent ions in the overall catalytic strategy employed by twister ribozymes. In this context, it is notable that twister retains wild-type activity when the phylogenetically conserved stem P1 is deleted, able to cleave a single nucleotide only. WIREs RNA 2017, 8:e1402. doi: 10.1002/wrna.1402 For further resources related to this article, please visit the WIREs website.
© 2016 The Authors. WIREs RNA published by Wiley Periodicals, Inc.
Figures



Similar articles
-
Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme.Proc Natl Acad Sci U S A. 2014 Sep 9;111(36):13028-33. doi: 10.1073/pnas.1414571111. Epub 2014 Aug 25. Proc Natl Acad Sci U S A. 2014. PMID: 25157168 Free PMC article.
-
Structural and Biochemical Properties of Novel Self-Cleaving Ribozymes.Molecules. 2017 Apr 24;22(4):678. doi: 10.3390/molecules22040678. Molecules. 2017. PMID: 28441772 Free PMC article. Review.
-
Ribozyme Catalysis with a Twist: Active State of the Twister Ribozyme in Solution Predicted from Molecular Simulation.J Am Chem Soc. 2016 Mar 9;138(9):3058-65. doi: 10.1021/jacs.5b12061. Epub 2016 Feb 25. J Am Chem Soc. 2016. PMID: 26859432 Free PMC article.
-
A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.Angew Chem Int Ed Engl. 2015 Dec 7;54(50):15128-15133. doi: 10.1002/anie.201506601. Epub 2015 Oct 16. Angew Chem Int Ed Engl. 2015. PMID: 26473980 Free PMC article.
-
Two distinct catalytic strategies in the hepatitis δ virus ribozyme cleavage reaction.Biochemistry. 2011 Nov 8;50(44):9424-33. doi: 10.1021/bi201157t. Epub 2011 Oct 17. Biochemistry. 2011. PMID: 22003985 Free PMC article. Review.
Cited by
-
Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme.Nat Commun. 2017 Oct 30;8(1):1180. doi: 10.1038/s41467-017-01276-y. Nat Commun. 2017. PMID: 29081514 Free PMC article.
-
The function of twister ribozyme variants in non-LTR retrotransposition in Schistosoma mansoni.Nucleic Acids Res. 2021 Oct 11;49(18):10573-10588. doi: 10.1093/nar/gkab818. Nucleic Acids Res. 2021. PMID: 34551436 Free PMC article.
-
Emergence of a "Cyclosome" in a Primitive Network Capable of Building "Infinite" Proteins.Life (Basel). 2019 Jun 18;9(2):51. doi: 10.3390/life9020051. Life (Basel). 2019. PMID: 31216720 Free PMC article.
-
Pseudoknot Formation Seeds the Twister Ribozyme Cleavage Reaction Coordinate.J Am Chem Soc. 2017 Jun 21;139(24):8186-8193. doi: 10.1021/jacs.7b01549. Epub 2017 Jun 9. J Am Chem Soc. 2017. PMID: 28598157 Free PMC article.
-
Mechanistic Debris Generated by Twister Ribozymes.ACS Chem Biol. 2017 Apr 21;12(4):886-891. doi: 10.1021/acschembio.7b00010. Epub 2017 Feb 21. ACS Chem Biol. 2017. PMID: 28191925 Free PMC article. Review.
References
-
- Lipkin D, Talbert PT, Cohn M. The mechanism of the alkaline hydrolysis of ribonucleic acids. Am Chem Soc 1954, 76:2871–2872.
-
- Bacher JE, Kauzmann W. The kinetics of hydrolysis of ribonucleic acid. J Am Chem Soc 1952, 74:3779–3786.
-
- Brown DM, Todd AR. Nucleotides. Part X: some observations on the structure and chemical behavior of the nucleic acids. J Chem Soc 1952, 52–58. doi: 10.1039/JR9520000052 - DOI
-
- Brown DM, Todd AR. The action of ribonuclease on simple esters of the monoribonucleotides. J Chem Soc 1953, 2040–2049. doi: 10.1039/JR9530002040 - DOI
-
- Brown DM, Magrath DI, Neilson AH, Todd AR. Hydrolysis of esters of monoribonucleotides. Nature 1956, 177:1124–1125.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

