Human Papillomavirus Drives Tumor Development Throughout the Head and Neck: Improved Prognosis Is Associated With an Immune Response Largely Restricted to the Oropharynx
- PMID: 27863190
- PMCID: PMC5477823
- DOI: 10.1200/JCO.2016.68.2955
Human Papillomavirus Drives Tumor Development Throughout the Head and Neck: Improved Prognosis Is Associated With an Immune Response Largely Restricted to the Oropharynx
Abstract
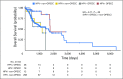
Purpose In squamous cell carcinomas of the head and neck (HNSCC), the increasing incidence of oropharyngeal squamous cell carcinomas (OPSCCs) is attributable to human papillomavirus (HPV) infection. Despite commonly presenting at late stage, HPV-driven OPSCCs are associated with improved prognosis compared with HPV-negative disease. HPV DNA is also detectable in nonoropharyngeal (non-OPSCC), but its pathogenic role and clinical significance are unclear. The objectives of this study were to determine whether HPV plays a causal role in non-OPSCC and to investigate whether HPV confers a survival benefit in these tumors. Methods Meta-analysis was used to build a cross-tissue gene-expression signature for HPV-driven cancer. Classifiers trained by machine-learning approaches were used to predict the HPV status of 520 HNSCCs profiled by The Cancer Genome Atlas project. DNA methylation data were similarly used to classify 464 HNSCCs and these analyses were integrated with genomic, histopathology, and survival data to permit a comprehensive comparison of HPV transcript-positive OPSCC and non-OPSCC. Results HPV-driven tumors accounted for 4.1% of non-OPSCCs. Regardless of anatomic site, HPV+ HNSCCs shared highly similar gene expression and DNA methylation profiles; nonkeratinizing, basaloid histopathological features; and lack of TP53 or CDKN2A alterations. Improved overall survival, however, was largely restricted to HPV-driven OPSCCs, which were associated with increased levels of tumor-infiltrating lymphocytes compared with HPV-driven non-OPSCCs. Conclusion Our analysis identified a causal role for HPV in transcript-positive non-OPSCCs throughout the head and neck. Notably, however, HPV-driven non-OPSCCs display a distinct immune microenvironment and clinical behavior compared with HPV-driven OPSCCs.
Conflict of interest statement
Authors’ disclosures of potential conflicts of interest are found in the article online at
Figures





References
-
- Doorbar J, Quint W, Banks L, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(suppl 5):F55–F70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

