Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control
- PMID: 27863240
- PMCID: PMC5125522
- DOI: 10.1016/j.cell.2016.10.024
Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control
Abstract
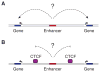
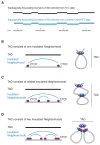
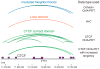
Understanding how transcriptional enhancers control over 20,000 protein-coding genes to maintain cell-type-specific gene expression programs in all human cells is a fundamental challenge in regulatory biology. Recent studies suggest that gene regulatory elements and their target genes generally occur within insulated neighborhoods, which are chromosomal loop structures formed by the interaction of two DNA sites bound by the CTCF protein and occupied by the cohesin complex. Here, we review evidence that insulated neighborhoods provide for specific enhancer-gene interactions, are essential for both normal gene activation and repression, form a chromosome scaffold that is largely preserved throughout development, and are perturbed by genetic and epigenetic factors in disease. Insulated neighborhoods are a powerful paradigm for gene control that provides new insights into development and disease.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

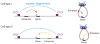


References
-
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
-
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
-
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

