A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility
- PMID: 27863247
- PMCID: PMC5131798
- DOI: 10.1016/j.cell.2016.10.043
A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility
Abstract
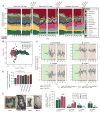
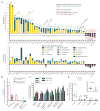
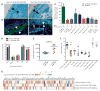
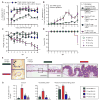
Despite the accepted health benefits of consuming dietary fiber, little is known about the mechanisms by which fiber deprivation impacts the gut microbiota and alters disease risk. Using a gnotobiotic mouse model, in which animals were colonized with a synthetic human gut microbiota composed of fully sequenced commensal bacteria, we elucidated the functional interactions between dietary fiber, the gut microbiota, and the colonic mucus barrier, which serves as a primary defense against enteric pathogens. We show that during chronic or intermittent dietary fiber deficiency, the gut microbiota resorts to host-secreted mucus glycoproteins as a nutrient source, leading to erosion of the colonic mucus barrier. Dietary fiber deprivation, together with a fiber-deprived, mucus-eroding microbiota, promotes greater epithelial access and lethal colitis by the mucosal pathogen, Citrobacter rodentium. Our work reveals intricate pathways linking diet, the gut microbiome, and intestinal barrier dysfunction, which could be exploited to improve health using dietary therapeutics.
Keywords: Akkermansia; Citrobacter rodentium; bacteroides; dietary fiber; gylcans; microbiome; microbiota; mucin; mucus layer; polysaccharides.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Veggies and Intact Grains a Day Keep the Pathogens Away.Cell. 2016 Nov 17;167(5):1161-1162. doi: 10.1016/j.cell.2016.10.047. Cell. 2016. PMID: 27863236
-
Gut microbiota: Low dietary fibre induces colonic mucus layer erosion by microbiota.Nat Rev Gastroenterol Hepatol. 2017 Jan;14(1):4. doi: 10.1038/nrgastro.2016.197. Epub 2016 Dec 7. Nat Rev Gastroenterol Hepatol. 2017. PMID: 27924078 No abstract available.
References
-
- Brownlee IA, Havler ME, Dettmar PW, Allen A, Pearson JP. Colonic mucus: secretion and turnover in relation to dietary fibre intake. Proc Nutr Soc. 2003;62:245–249. - PubMed
-
- Burkitt DP, Walker ARP, Painter NS. Effect of dietary fibre on stools and transit-times, and its role in the causation of disease. Lancet. 1972;300:1408–1411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

