Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells
- PMID: 27863251
- PMCID: PMC5119954
- DOI: 10.1016/j.cell.2016.10.026
Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells
Abstract
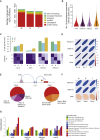
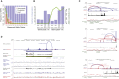
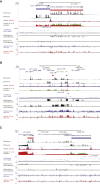
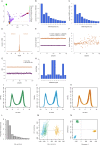
Characterizing the multifaceted contribution of genetic and epigenetic factors to disease phenotypes is a major challenge in human genetics and medicine. We carried out high-resolution genetic, epigenetic, and transcriptomic profiling in three major human immune cell types (CD14+ monocytes, CD16+ neutrophils, and naive CD4+ T cells) from up to 197 individuals. We assess, quantitatively, the relative contribution of cis-genetic and epigenetic factors to transcription and evaluate their impact as potential sources of confounding in epigenome-wide association studies. Further, we characterize highly coordinated genetic effects on gene expression, methylation, and histone variation through quantitative trait locus (QTL) mapping and allele-specific (AS) analyses. Finally, we demonstrate colocalization of molecular trait QTLs at 345 unique immune disease loci. This expansive, high-resolution atlas of multi-omics changes yields insights into cell-type-specific correlation between diverse genomic inputs, more generalizable correlations between these inputs, and defines molecular events that may underpin complex disease risk.
Keywords: DNA methylation; EWAS; QTL; allele specific; histone modification; immune; monocyte; neutrophil; t-cell; transription.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures










Comment in
-
Concerted Genetic Function in Blood Traits.Cell. 2016 Nov 17;167(5):1167-1169. doi: 10.1016/j.cell.2016.10.055. Cell. 2016. PMID: 27863238
References
-
- Abnizova I., Skelly T., Naumenko F., Whiteford N., Brown C., Cox T. Statistical comparison of methods to estimate the error probability in short-read Illumina sequencing. J. Bioinform. Comput. Biol. 2010;8:579–591. - PubMed
-
- Allum F., Shao X., Guénard F., Simon M.M., Busche S., Caron M., Lambourne J., Lessard J., Tandre K., Hedman A.K., Multiple Tissue Human Expression Resource Consortium Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat. Commun. 2015;6:7211. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

