Aberrant overexpression of ADAR1 promotes gastric cancer progression by activating mTOR/p70S6K signaling
- PMID: 27863387
- PMCID: PMC5349904
- DOI: 10.18632/oncotarget.13354
Aberrant overexpression of ADAR1 promotes gastric cancer progression by activating mTOR/p70S6K signaling
Abstract
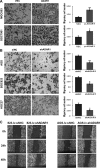
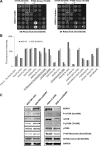
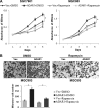
ADAR1, one of adenosine deaminases acting on RNA, modulates RNA transcripts through converting adenosine (A) to inosine (I) by deamination. Emerging evidence has implicated that ADAR1 plays an important role in a few of human cancers, however, its expression and physiological significance in gastric cancer remain undefined. In the present study, we demonstrated that ADAR1 was frequently overexpressed in gastric cancer samples by quantitative real-time PCR analysis. In a gastric cancer tissue microarray, ADAR1 staining was closely correlated with tumor stage (P < 0.001) and N classification (P < 0.001). Functional analysis indicated that ADAR1 overexpression promoted cell proliferation and migration in vitro, whereas ADAR1 knockdown resulted in an opposite phenotypes. Furthermore, ADAR1 knockdown also inhibited tumorigenicity and lung metastasis potential of gastric cancer cells in nude mice models. Mechanistically, ADAR1 expression had a significant effect on phosphorylation level of mTOR, p70S kinase, and S6 ribosomal protein, implying its involvement in the regulation of mTOR signaling pathway. We conclude that ADAR1 contributes to gastric cancer development and progression via activating mTOR/p70S6K/S6 ribosomal protein signaling axis. Our findings suggest that ADAR1 may be a valuable biomarker for GC diagnosis and prognosis and may represent a new novel therapeutic opportunities.
Keywords: ADAR1; gastric cancer; mTOR; metastasis; tumorigenecity.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




Similar articles
-
Rab1A knockdown represses proliferation and promotes apoptosis in gastric cancer cells by inhibition of mTOR/p70S6K pathway.Arch Biochem Biophys. 2020 May 30;685:108352. doi: 10.1016/j.abb.2020.108352. Epub 2020 Mar 30. Arch Biochem Biophys. 2020. PMID: 32240637
-
The role of mTOR and phospho-p70S6K in pathogenesis and progression of gastric carcinomas: an immunohistochemical study on tissue microarray.J Exp Clin Cancer Res. 2009 Dec 13;28(1):152. doi: 10.1186/1756-9966-28-152. J Exp Clin Cancer Res. 2009. PMID: 20003385 Free PMC article.
-
Enhanced AZIN1 RNA editing and overexpression of its regulatory enzyme ADAR1 are important prognostic biomarkers in gastric cancer.J Transl Med. 2018 Dec 18;16(1):366. doi: 10.1186/s12967-018-1740-z. J Transl Med. 2018. PMID: 30563560 Free PMC article.
-
STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy.Mol Cancer. 2018 Jan 12;17(1):6. doi: 10.1186/s12943-017-0756-y. Mol Cancer. 2018. PMID: 29329543 Free PMC article.
-
ADAR1 promotes the epithelial-to-mesenchymal transition and stem-like cell phenotype of oral cancer by facilitating oncogenic microRNA maturation.J Exp Clin Cancer Res. 2019 Jul 17;38(1):315. doi: 10.1186/s13046-019-1300-2. J Exp Clin Cancer Res. 2019. PMID: 31315644 Free PMC article.
Cited by
-
ADAR1 may be involved in the proliferation of acute myeloid leukemia cells via regulation of the Wnt pathway.Cancer Manag Res. 2019 Sep 20;11:8547-8555. doi: 10.2147/CMAR.S210504. eCollection 2019. Cancer Manag Res. 2019. PMID: 31572009 Free PMC article.
-
Targeting ADAR1 with a small molecule for the treatment of prostate cancer.Nat Cancer. 2025 Mar;6(3):474-492. doi: 10.1038/s43018-025-00907-4. Epub 2025 Feb 10. Nat Cancer. 2025. PMID: 39930013
-
ADAR1 Promotes the Progression and Temozolomide Resistance of Glioma Through p62-Mediated Selective Autophagy.CNS Neurosci Ther. 2025 Jan;31(1):e70168. doi: 10.1111/cns.70168. CNS Neurosci Ther. 2025. PMID: 39825637 Free PMC article.
-
Adenosine-to-inosine RNA editing in cancer: molecular mechanisms and downstream targets.Protein Cell. 2025 Jun 20;16(6):391-417. doi: 10.1093/procel/pwae039. Protein Cell. 2025. PMID: 39126156 Free PMC article. Review.
-
Cancer metastasis under the magnifying glass of epigenetics and epitranscriptomics.Cancer Metastasis Rev. 2023 Dec;42(4):1071-1112. doi: 10.1007/s10555-023-10120-3. Epub 2023 Jun 28. Cancer Metastasis Rev. 2023. PMID: 37369946 Free PMC article. Review.
References
-
- Soundararajan R, Stearns TM, Griswold AL, Mehta A, Czachor A, Fukumoto J, Lockey RF, King BL, Kolliputi N. Detection of canonical A-to-G editing events at 3′ UTRs and microRNA target sites in human lungs using next-generation sequencing. Oncotarget. 2015;6:35726–35736. doi: 10.18632/oncotarget.6132. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

