Different non-synonymous polymorphisms modulate the interaction of the WRN protein to its protein partners and its enzymatic activities
- PMID: 27863399
- PMCID: PMC5349866
- DOI: 10.18632/oncotarget.13341
Different non-synonymous polymorphisms modulate the interaction of the WRN protein to its protein partners and its enzymatic activities
Abstract
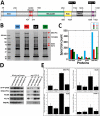
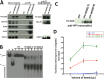
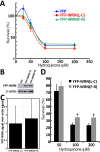
Werner syndrome (WS) is characterized by the premature onset of several age-associated pathologies including cancer. The protein defective in WS patients (WRN) is a helicase/exonuclease involved in DNA replication and repair. Here, we present the results of a large-scale proteome analysis that has been undertaken to determine protein partners of different polymorphic WRN proteins found with relatively high prevalence in the human population. We expressed different fluorescently tagged-WRN (eYFP-WRN) variants in human 293 embryonic kidney cells (HEK293) and used a combination of affinity-purification and mass spectrometry to identify different compositions of WRN-associated protein complexes. We found that a WRN variant containing a phenylalanine residue at position 1074 and an arginine at position 1367 (eYFP-WRN(F-R)) possesses more affinity for DNA-PKc, KU86, KU70, and PARP1 than a variant containing a leucine at position 1074 and a cysteine at position 1367 (eYFP-WRN(L-C)). Such results were confirmed in a WRN-deficient background using WS fibroblasts. Interestingly, the exonuclase activity of WRN recovered from immunoprecipitated eYFP-WRN(L-C) variant was lower than the eYFP-WRN(F-R) in WS cells. Finally, HEK293 cells and WS fibroblasts overexpressing the eYFP-WRN(F-R) variant were more resistant to the benzene metabolite hydroquinone than cells expressing the eYFP-WRN(L-C) variant. These results indicate that the protein-protein interaction landscape of WRN is subject to modulation by polymorphic amino acids, a characteristic associated with distinctive cell survival outcome.
Keywords: Gerotarget; exonuclease; mass spectrometry; polymorphism; proteomics; werner syndrome.
Conflict of interest statement
None to declare.
Figures



References
-
- Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. - PubMed
-
- Salk D. Werner's syndrome: a review of recent research with an analysis of connective tissue metabolism, growth control of cultured cells, and chromosomal aberrations. Hum Genet. 1982;62:1–5. - PubMed
-
- Ozgenc A, Loeb LA. Current advances in unraveling the function of the Werner syndrome protein. Mutat Res. 2005;577:237–51. - PubMed
-
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–62. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

