Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers
- PMID: 27863400
- PMCID: PMC5352059
- DOI: 10.18632/oncotarget.13346
Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers
Abstract
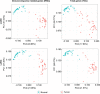
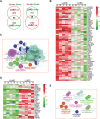
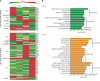
Lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) are the two major subtypes of lung cancer, with LUSC exhibits faster progression rate than LUAD. To investigate the roles of immune-response related genes (IRGs) in lung cancer progression, we used LUAD and LUSC samples at different cancer progression stages, and identified that the expression profiles of IRGs could serve as a better classification marker for cancerous tissues of both LUAD and LUSC. We found that the expression changes of IRGs were different between LUAD and LUSC. Cell cycle promoting genes, including KIFs and proteasomes, showed faster up-regulation in LUSC, whereas immune response promoting genes, including MHC molecules and chemokines, were more rapidly repressed in LUSC. Comparative pathway analysis revealed that members of the Toll-like receptor and T cell receptor signaling pathways exhibited diverged expression changes between LUAD and LUSC, especially at the early cancer stages. Our results revealed the difference of LUAD and LUSC from the immune response point of view, and provided new clues for the differential treatment of LUAD and LUSC.
Keywords: LUAD; LUSC; cancer progression rate; expression features; immune-response related genes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. - PubMed
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115–132. - PubMed
-
- Detterbeck FC, Boffa DJ, Tanoue LT. The New Lung Cancer Staging System. Chest. 2009;136:260–271. - PubMed
-
- Subramanian J, Govindan R. Lung cancer in never smokers: a review. Journal of clinical oncology. 2007;25:561–570. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

