Microarray analysis reveals a potential role of LncRNAs expression in cardiac cell proliferation
- PMID: 27863467
- PMCID: PMC5116129
- DOI: 10.1186/s12861-016-0139-4
Microarray analysis reveals a potential role of LncRNAs expression in cardiac cell proliferation
Abstract
Background: Long non-coding RNAs (LncRNAs) have been identified to play important roles in epigenetic processes that underpin organogenesis. However, the role of LncRNAs in the regulation of transition from fetal to adult life of human heart has not been evaluated.
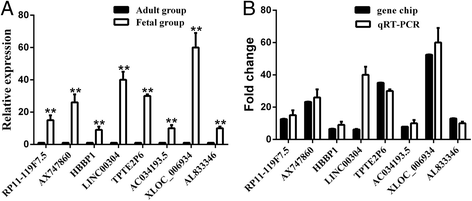
Methods: Immunofiuorescent staining was used to determine the extent of cardiac cell proliferation. Human LncRNA microarrays were applied to define gene expression signatures of the fetal (13-17 weeks of gestation, n = 4) and adult hearts (30-40 years old, n = 4). Pathway analysis was performed to predict the function of differentially expressed mRNAs (DEM). DEM related to cell proliferation were selected to construct a lncRNA-mRNA co-expression network. Eight lncRNAs were confirmed by quantificational real-time polymerase chain reaction (n = 6).
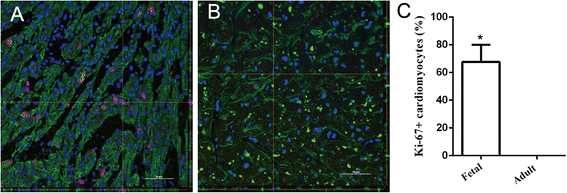
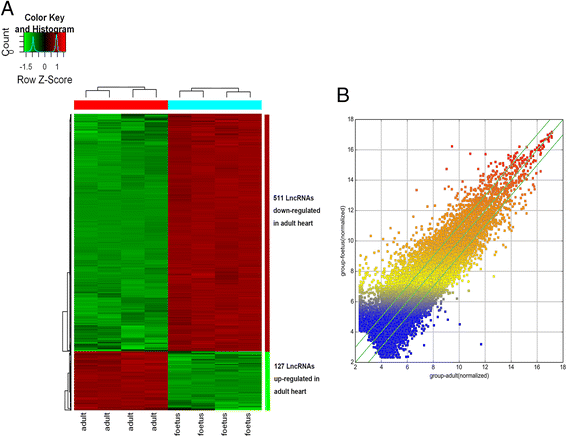
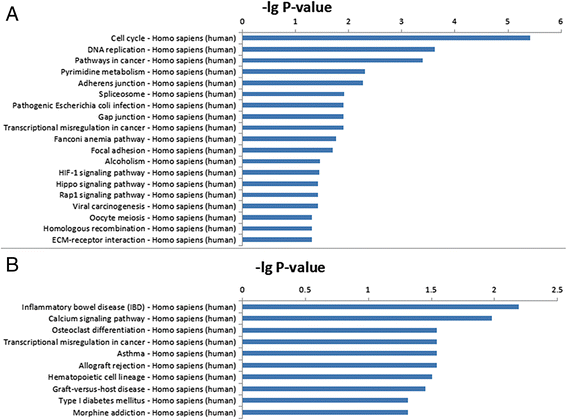
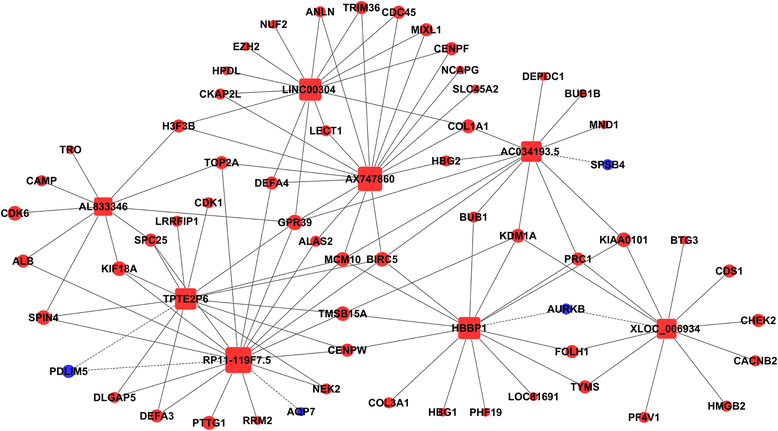
Results: Cardiac cell proliferation was significant in the fetal heart. Two thousand six hundred six lncRNAs and 3079 mRNAs were found to be differentially expressed. Cell cycle was the most enriched pathway in down-regulated genes in the adult heart. Eight lncRNAs (RP11-119 F7.5, AX747860, HBBP1, LINC00304, TPTE2P6, AC034193.5, XLOC_006934 and AL833346) were predicted to play a central role in cardiac cell proliferation.
Conclusions: We discovered a profile of lncRNAs differentially expressed between the human fetal and adult heart. Several meaningful lncRNAs involved in cardiac cell proliferation were disclosed.
Keywords: Cardiac cell proliferation; Human heart; Long non-coding RNA; Microarray.
Figures
Similar articles
-
The long non-coding road to endogenous cardiac regeneration.Heart Fail Rev. 2019 Jul;24(4):587-600. doi: 10.1007/s10741-019-09782-5. Heart Fail Rev. 2019. PMID: 30900115 Review.
-
Long Non-coding RNA-mRNA Correlation Analysis Reveals the Potential Role of HOTAIR in Pathogenesis of Sporadic Thoracic Aortic Aneurysm.Eur J Vasc Endovasc Surg. 2017 Sep;54(3):303-314. doi: 10.1016/j.ejvs.2017.06.010. Epub 2017 Jul 27. Eur J Vasc Endovasc Surg. 2017. PMID: 28757056
-
Differential Expression of Long Noncoding RNAs During Cardiac Allograft Rejection.Transplantation. 2017 Jan;101(1):83-91. doi: 10.1097/TP.0000000000001463. Transplantation. 2017. PMID: 27575691
-
The long noncoding RNA expression profiles of paroxysmal atrial fibrillation identified by microarray analysis.Gene. 2018 Feb 5;642:125-134. doi: 10.1016/j.gene.2017.11.025. Epub 2017 Nov 10. Gene. 2018. PMID: 29129807
-
Aberrant expression of long noncoding RNAs in cumulus cells isolated from PCOS patients.J Assist Reprod Genet. 2016 Jan;33(1):111-21. doi: 10.1007/s10815-015-0630-z. Epub 2015 Dec 9. J Assist Reprod Genet. 2016. PMID: 26650608 Free PMC article.
Cited by
-
Gene regulation of mammalian long non-coding RNA.Mol Genet Genomics. 2018 Feb;293(1):1-15. doi: 10.1007/s00438-017-1370-9. Epub 2017 Sep 11. Mol Genet Genomics. 2018. PMID: 28894972 Review.
-
The Function and Therapeutic Potential of Long Non-coding RNAs in Cardiovascular Development and Disease.Mol Ther Nucleic Acids. 2017 Sep 15;8:494-507. doi: 10.1016/j.omtn.2017.07.014. Epub 2017 Jul 28. Mol Ther Nucleic Acids. 2017. PMID: 28918050 Free PMC article. Review.
-
LINC00365-SCGB2A1 axis inhibits the viability of breast cancer through targeting NF-κB signaling.Oncol Lett. 2020 Jan;19(1):753-762. doi: 10.3892/ol.2019.11166. Epub 2019 Nov 29. Oncol Lett. 2020. PMID: 31897191 Free PMC article.
-
The roles of long noncoding RNAs in myocardial pathophysiology.Biosci Rep. 2019 Nov 29;39(11):BSR20190966. doi: 10.1042/BSR20190966. Biosci Rep. 2019. PMID: 31694052 Free PMC article. Review.
-
The long non-coding road to endogenous cardiac regeneration.Heart Fail Rev. 2019 Jul;24(4):587-600. doi: 10.1007/s10741-019-09782-5. Heart Fail Rev. 2019. PMID: 30900115 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

