A genomic case study of desmoplastic small round cell tumor: comprehensive analysis reveals insights into potential therapeutic targets and development of a monitoring tool for a rare and aggressive disease
- PMID: 27863505
- PMCID: PMC5116179
- DOI: 10.1186/s40246-016-0092-0
A genomic case study of desmoplastic small round cell tumor: comprehensive analysis reveals insights into potential therapeutic targets and development of a monitoring tool for a rare and aggressive disease
Abstract
Background: Genome-wide profiling of rare tumors is crucial for improvement of diagnosis, treatment, and, consequently, achieving better outcomes. Desmoplastic small round cell tumor (DSRCT) is a rare type of sarcoma arising from mesenchymal cells of abdominal peritoneum that usually develops in male adolescents and young adults. A specific translocation, t(11;22)(p13;q12), resulting in EWS and WT1 gene fusion is the only recurrent molecular hallmark and no other genetic factor has been associated to this aggressive tumor. Here, we present a comprehensive genomic profiling of one DSRCT affecting a 26-year-old male, who achieved an excellent outcome.
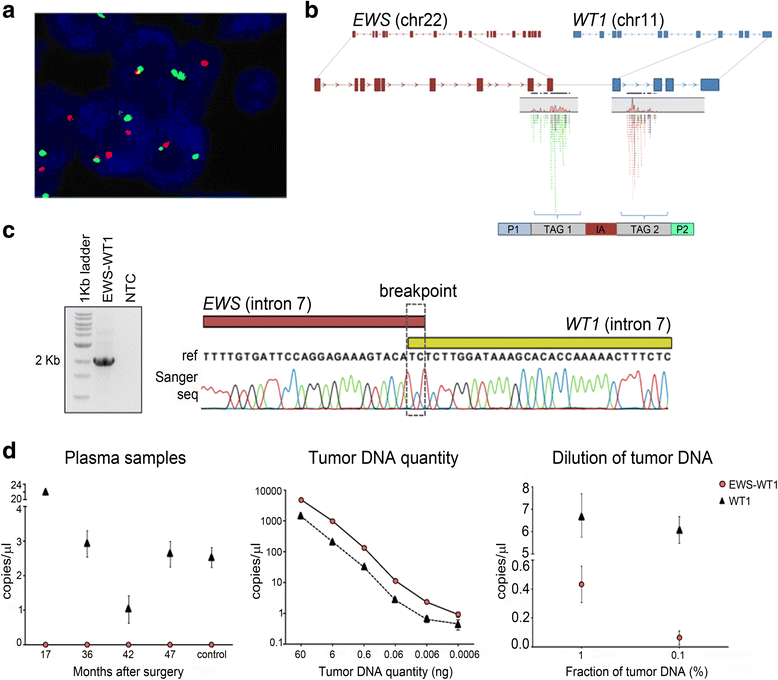
Methods: We investigated somatic and germline variants through whole-exome sequencing using a family based approach and, by array CGH, we explored the occurrence of genomic imbalances. Additionally, we performed mate-paired whole-genome sequencing for defining the specific breakpoint of the EWS-WT1 translocation, allowing us to develop a personalized tumor marker for monitoring the patient by liquid biopsy.
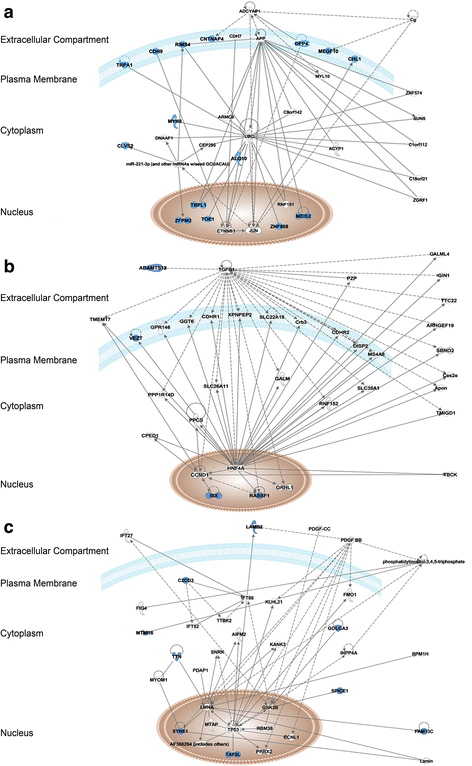
Results: We identified genetic variants leading to protein alterations including 12 somatic and 14 germline events (11 germline compound heterozygous mutations and 3 rare homozygous polymorphisms) affecting genes predominantly involved in mesenchymal cell differentiation pathways. Regarding copy number alterations (CNA) few events were detected, mainly restricted to gains in chromosomes 5 and 18 and losses at 11p, 13q, and 22q. The deletions at 11p and 22q indicated the presence of the classic translocation, t(11;22)(p13;q12). In addition, the mapping of the specific genomic breakpoint of the EWS-WT1 gene fusion allowed the design of a personalized biomarker for assessing circulating tumor DNA (ctDNA) in plasma during patient follow-up. This biomarker has been used in four post-treatment blood samples, 3 years after surgery, and no trace of EWS-WT1 gene fusion was detected, in accordance with imaging tests showing no evidence of disease and with the good general health status of the patient.
Conclusions: Overall, our findings revealed genes with potential to be associated with risk assessment and tumorigenesis of this rare type of sarcoma. Additionally, we established a liquid biopsy approach for monitoring patient follow-up based on genomic information that can be similarly adopted for patients diagnosed with a rare tumor.
Keywords: Desmoplastic small round cell tumor; EWS-WT1 gene fusion; Genomic profiling; Liquid biopsy; Personalized biomarker; Whole-exome sequencing.
Figures

Similar articles
-
Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): desmoplastic small round-cell tumor and its variants.J Clin Oncol. 1998 Sep;16(9):3028-36. doi: 10.1200/JCO.1998.16.9.3028. J Clin Oncol. 1998. PMID: 9738572
-
Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor.Cancer Res. 1994 Jun 1;54(11):2837-40. Cancer Res. 1994. PMID: 8187063
-
Characterization of the genomic breakpoint and chimeric transcripts in the EWS-WT1 gene fusion of desmoplastic small round cell tumor.Proc Natl Acad Sci U S A. 1995 Feb 14;92(4):1028-32. doi: 10.1073/pnas.92.4.1028. Proc Natl Acad Sci U S A. 1995. PMID: 7862627 Free PMC article.
-
Fusion of the EWS1 and WT1 genes as a result of the t(11;22)(p13;q12) translocation in desmoplastic small round cell tumors.Med Pediatr Oncol. 1996 Nov;27(5):434-9. doi: 10.1002/(SICI)1096-911X(199611)27:5<434::AID-MPO8>3.0.CO;2-N. Med Pediatr Oncol. 1996. PMID: 8827070 Review.
-
Updates on the multimodality management of desmoplastic small round cell tumor.J Surg Oncol. 2012 May;105(6):617-21. doi: 10.1002/jso.22130. Epub 2012 Jan 3. J Surg Oncol. 2012. PMID: 22215508 Review.
Cited by
-
Small round cell sarcomas.Nat Rev Dis Primers. 2022 Oct 6;8(1):66. doi: 10.1038/s41572-022-00393-3. Nat Rev Dis Primers. 2022. PMID: 36202860 Review.
-
Genomic Breakpoint Characterization and Transcriptome Analysis of Metastatic, Recurrent Desmoplastic Small Round Cell Tumor.Sarcoma. 2023 Jul 6;2023:6686702. doi: 10.1155/2023/6686702. eCollection 2023. Sarcoma. 2023. PMID: 37457440 Free PMC article.
-
Liquid Biopsy for Spinal Tumors: On the Frontiers of Clinical Application.Global Spine J. 2025 Jan;15(1_suppl):16S-28S. doi: 10.1177/21925682231222012. Global Spine J. 2025. PMID: 39801114 Free PMC article.
-
Changing incidence and survival of desmoplastic small round cell tumor in the USA.Proc (Bayl Univ Med Cent). 2022 Mar 22;35(4):415-419. doi: 10.1080/08998280.2022.2049581. eCollection 2022. Proc (Bayl Univ Med Cent). 2022. PMID: 35754588 Free PMC article.
-
Mini-Review on Targeted Treatment of Desmoplastic Small Round Cell Tumor.Front Oncol. 2020 Apr 21;10:518. doi: 10.3389/fonc.2020.00518. eCollection 2020. Front Oncol. 2020. PMID: 32373525 Free PMC article. Review.
References
-
- Liu J, Nau MM, Yeh JC, Allegra CJ, Chu E, Wright JJ. Molecular heterogeneity and function of EWS-WT1 fusion transcripts in desmoplastic small round cell tumors. Clin Cancer Res. 2000;6:3522–3529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

