The BLUEPRINT Data Analysis Portal
- PMID: 27863955
- PMCID: PMC5919098
- DOI: 10.1016/j.cels.2016.10.021
The BLUEPRINT Data Analysis Portal
Abstract
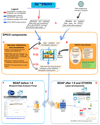
The impact of large and complex epigenomic datasets on biological insights or clinical applications is limited by the lack of accessibility by easy, intuitive, and fast tools. Here, we describe an epigenomics comparative cyber-infrastructure (EPICO), an open-access reference set of libraries to develop comparative epigenomic data portals. Using EPICO, large epigenome projects can make available their rich datasets to the community without requiring specific technical skills. As a first instance of EPICO, we implemented the BLUEPRINT Data Analysis Portal (BDAP). BDAP provides a desktop for the comparative analysis of epigenomes of hematopoietic cell types based on results, such as the position of epigenetic features, from basic analysis pipelines. The BDAP interface facilitates interactive exploration of genomic regions, genes, and pathways in the context of differentiation of hematopoietic lineages. This work represents initial steps toward broadly accessible integrative analysis of epigenomic data across international consortia. EPICO can be accessed at https://github.com/inab, and BDAP can be accessed at http://blueprint-data.bsc.es.
Keywords: BLUEPRINT Data Analysis Portal; BLUEPRINT epigenomes; International Human Epigenomes Consortium; bioinformatics; cyber-infrastructure; epigenomic data mining tools; epigenomic data visualization; epigenomics; hematopoiesis; leukemia.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
References
-
- Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, Caricasole A, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30(3):224–226. - PubMed
-
- BP-analysis. BLUEPRINT Analysis descriptions release 20160816. 2016 ftp://ftp.ebi.ac.uk/pub/databases/blueprint/releases/20160816/homo_sapiens/
-
- BP-Data_analysis. BLUEPRINT Data Analysis Portal GitHub repository. 2016 https://github.com/inab/epico-data-analysis-portal.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

