Robust skull stripping using multiple MR image contrasts insensitive to pathology
- PMID: 27864083
- PMCID: PMC5321800
- DOI: 10.1016/j.neuroimage.2016.11.017
Robust skull stripping using multiple MR image contrasts insensitive to pathology
Abstract
Automatic skull-stripping or brain extraction of magnetic resonance (MR) images is often a fundamental step in many neuroimage processing pipelines. The accuracy of subsequent image processing relies on the accuracy of the skull-stripping. Although many automated stripping methods have been proposed in the past, it is still an active area of research particularly in the context of brain pathology. Most stripping methods are validated on T1-w MR images of normal brains, especially because high resolution T1-w sequences are widely acquired and ground truth manual brain mask segmentations are publicly available for normal brains. However, different MR acquisition protocols can provide complementary information about the brain tissues, which can be exploited for better distinction between brain, cerebrospinal fluid, and unwanted tissues such as skull, dura, marrow, or fat. This is especially true in the presence of pathology, where hemorrhages or other types of lesions can have similar intensities as skull in a T1-w image. In this paper, we propose a sparse patch based Multi-cONtrast brain STRipping method (MONSTR),2 where non-local patch information from one or more atlases, which contain multiple MR sequences and reference delineations of brain masks, are combined to generate a target brain mask. We compared MONSTR with four state-of-the-art, publicly available methods: BEaST, SPECTRE, ROBEX, and OptiBET. We evaluated the performance of these methods on 6 datasets consisting of both healthy subjects and patients with various pathologies. Three datasets (ADNI, MRBrainS, NAMIC) are publicly available, consisting of 44 healthy volunteers and 10 patients with schizophrenia. Other three in-house datasets, comprising 87 subjects in total, consisted of patients with mild to severe traumatic brain injury, brain tumors, and various movement disorders. A combination of T1-w, T2-w were used to skull-strip these datasets. We show significant improvement in stripping over the competing methods on both healthy and pathological brains. We also show that our multi-contrast framework is robust and maintains accurate performance across different types of acquisitions and scanners, even when using normal brains as atlases to strip pathological brains, demonstrating that our algorithm is applicable even when reference segmentations of pathological brains are not available to be used as atlases.
Keywords: Atlas; Brain extraction; Non-local; Patches; Segmentation; Skull stripping; Sparsity.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
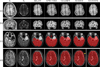
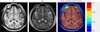
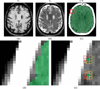





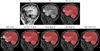
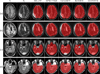





References
-
- Boesen K, Rehm K, Schaper K, Stoltzner S, Woods R, Lüders E, Rottenberg D. Quantitative comparison of four brain extraction algorithms. NeuroImage. 2004;22(3):1255–1261. - PubMed
-
- Buades A, Coll B, Morel JM. A Non-Local Algorithm for Image Denoising. Intl. Conf. on Comp. Vision. and Patt. Recog. (CVPR) 2005;2:60–65.
-
- Burgos N, Cardoso MJ, Thielemans K, Modat M, Pedemonte S, Dickson J, Barnes A, Ahmed R, Mahoney CJ, Schott JM, Duncan JS, Atkinson D, Arridge SR, Hutton BF, Ourselin S. Attenuation correction synthesis for hybrid PET-MR scanners: Application to brain studies. IEEE Trans. Med. Imag. 2014;33(12):2332–2341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

