Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes
- PMID: 27864352
- PMCID: PMC5287227
- DOI: 10.1101/gr.212720.116
Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes
Abstract
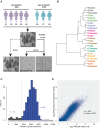
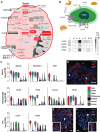
Blood glucose levels are tightly controlled by the coordinated action of at least four cell types constituting pancreatic islets. Changes in the proportion and/or function of these cells are associated with genetic and molecular pathophysiology of monogenic, type 1, and type 2 (T2D) diabetes. Cellular heterogeneity impedes precise understanding of the molecular components of each islet cell type that govern islet (dys)function, particularly the less abundant delta and gamma/pancreatic polypeptide (PP) cells. Here, we report single-cell transcriptomes for 638 cells from nondiabetic (ND) and T2D human islet samples. Analyses of ND single-cell transcriptomes identified distinct alpha, beta, delta, and PP/gamma cell-type signatures. Genes linked to rare and common forms of islet dysfunction and diabetes were expressed in the delta and PP/gamma cell types. Moreover, this study revealed that delta cells specifically express receptors that receive and coordinate systemic cues from the leptin, ghrelin, and dopamine signaling pathways implicating them as integrators of central and peripheral metabolic signals into the pancreatic islet. Finally, single-cell transcriptome profiling revealed genes differentially regulated between T2D and ND alpha, beta, and delta cells that were undetectable in paired whole islet analyses. This study thus identifies fundamental cell-type-specific features of pancreatic islet (dys)function and provides a critical resource for comprehensive understanding of islet biology and diabetes pathogenesis.
© 2017 Lawlor et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Abdallah BM, Ditzel N, Laborda J, Karsenty G, Kassem M. 2015. DLK1 regulates whole-body glucose metabolism: a negative feedback regulation of the osteocalcin-insulin loop. Diabetes 64: 3069–3080. - PubMed
-
- Andersson SA, Olsson AH, Esguerra JL, Heimann E, Ladenvall C, Edlund A, Salehi A, Taneera J, Degerman E, Groop L, et al. 2012. Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes. Mol Cell Endocrinol 364: 36–45. - PubMed
-
- Bader E, Migliorini A, Gegg M, Moruzzi N, Gerdes J, Roscioni SS, Bakhti M, Brandl E, Irmler M, Beckers J, et al. 2016. Identification of proliferative and mature β-cells in the islets of Langerhans. Nature 535: 430–434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
