Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs
- PMID: 27864467
- PMCID: PMC5154947
- DOI: 10.1084/jem.20161135
Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs
Abstract
In humans, conventional dendritic cells (cDCs) exist as two unique populations characterized by expression of CD1c and CD141. cDCs arise from increasingly restricted but well-defined bone marrow progenitors that include the common DC progenitor that differentiates into the pre-cDC, which is the direct precursor of cDCs. In this study, we show that pre-cDCs in humans are heterogeneous, consisting of two distinct populations of precursors that are precommitted to become either CD1c+ or CD141+ cDCs. The two groups of lineage-primed precursors can be distinguished based on differential expression of CD172a. Both subpopulations of pre-cDCs arise in the adult bone marrow and can be found in cord blood and adult peripheral blood. Gene expression analysis revealed that CD172a+ and CD172a- pre-cDCs represent developmentally discrete populations that differentially express lineage-restricted transcription factors. A clinical trial of Flt3L injection revealed that this cytokine increases the number of both CD172a- and CD172a+ pre-cDCs in human peripheral blood.
© 2016 Breton et al.
Figures
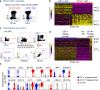

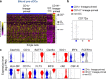

References
-
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W., et al. . 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281. 10.1084/jem.20100348 - DOI - PMC - PubMed
-
- Cohn L., Chatterjee B., Esselborn F., Smed-Sörensen A., Nakamura N., Chalouni C., Lee B.C., Vandlen R., Keler T., Lauer P., et al. . 2013. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J. Exp. Med. 210:1049–1063. 10.1084/jem.20121251 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

