Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways
- PMID: 27864472
- PMCID: PMC5238794
- DOI: 10.1261/rna.059055.116
Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways
Abstract
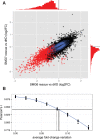
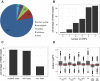
Besides degrading aberrant mRNAs that harbor a premature translation termination codon (PTC), nonsense-mediated mRNA decay (NMD) also targets many seemingly "normal" mRNAs that encode for full-length proteins. To identify a bona fide set of such endogenous NMD targets in human cells, we applied a meta-analysis approach in which we combined transcriptome profiling of knockdowns and rescues of the three NMD factors UPF1, SMG6, and SMG7. We provide evidence that this combinatorial approach identifies NMD-targeted transcripts more reliably than previous attempts that focused on inactivation of single NMD factors. Our data revealed that SMG6 and SMG7 act on essentially the same transcripts, indicating extensive redundancy between the endo- and exonucleolytic decay routes. Besides mRNAs, we also identified as NMD targets many long noncoding RNAs as well as miRNA and snoRNA host genes. The NMD target feature with the most predictive value is an intron in the 3' UTR, followed by the presence of upstream open reading frames (uORFs) and long 3' UTRs. Furthermore, the 3' UTRs of NMD-targeted transcripts tend to have an increased GC content and to be phylogenetically less conserved when compared to 3' UTRs of NMD insensitive transcripts.
Keywords: RNA turnover; SMG6; SMG7; UPF1; bioinformatics analysis; mRNA-seq; nonsense-mediated mRNA decay; post-transcriptional gene regulation.
© 2017 Colombo et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. - PubMed
-
- Balistreri G, Horvath P, Schweingruber C, Zünd D, McInerney G, Merits A, Mühlemann O, Azzalin C, Helenius A. 2014. The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 16: 403–411. - PubMed
-
- Boehm V, Haberman N, Ottens F, Ule J, Gehring NH. 2014. 3′ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep 9: 555–568. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
