Protecting from Envelope Stress: Variations on the Phage-Shock-Protein Theme
- PMID: 27865622
- PMCID: PMC5551406
- DOI: 10.1016/j.tim.2016.10.001
Protecting from Envelope Stress: Variations on the Phage-Shock-Protein Theme
Erratum in
-
Protecting from Envelope Stress: Variations on the Phage-Shock-Protein Theme: (Trends in Microbiology 25, 205-216; 2017).Trends Microbiol. 2017 Mar;25(3):242. doi: 10.1016/j.tim.2016.11.010. Epub 2016 Dec 4. Trends Microbiol. 2017. PMID: 27923541 No abstract available.
Abstract
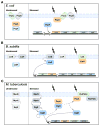
During envelope stress, critical inner-membrane functions are preserved by the phage-shock-protein (Psp) system, a stress response that emerged from work with Escherichia coli and other Gram-negative bacteria. Reciprocal regulatory interactions and multiple effector functions are well documented in these organisms. Searches for the Psp system across phyla reveal conservation of only one protein, PspA. However, examination of Firmicutes and Actinobacteria reveals that PspA orthologs associate with non-orthologous regulatory and effector proteins retaining functions similar to those in Gram-negative counterparts. Conservation across phyla emphasizes the long-standing importance of the Psp system in prokaryotes, while inter- and intra-phyla variations within the system indicate adaptation to different cell envelope structures, bacterial lifestyles, and/or bacterial morphogenetic strategies.
Keywords: Actinobacteria; ClgR; Firmicutes; Lia; PspA; envelope stress response; morphogenesis.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

References
-
- Rowley G, et al. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4(5):383–94. - PubMed
-
- Manganelli R, Provvedi R. An integrated regulatory network including two positive feedback loops to modulate the activity of sigma(E) in mycobacteria. Mol Microbiol. 2010;75(3):538–42. - PubMed
-
- Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev. 2008;32(1):107–46. - PubMed
-
- Darwin AJ. The phage-shock-protein response. Mol Microbiol. 2005;57(3):621–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

