Colocalization of GWAS and eQTL Signals Detects Target Genes
- PMID: 27866706
- PMCID: PMC5142122
- DOI: 10.1016/j.ajhg.2016.10.003
Colocalization of GWAS and eQTL Signals Detects Target Genes
Abstract
The vast majority of genome-wide association study (GWAS) risk loci fall in non-coding regions of the genome. One possible hypothesis is that these GWAS risk loci alter the individual's disease risk through their effect on gene expression in different tissues. In order to understand the mechanisms driving a GWAS risk locus, it is helpful to determine which gene is affected in specific tissue types. For example, the relevant gene and tissue could play a role in the disease mechanism if the same variant responsible for a GWAS locus also affects gene expression. Identifying whether or not the same variant is causal in both GWASs and expression quantitative trail locus (eQTL) studies is challenging because of the uncertainty induced by linkage disequilibrium and the fact that some loci harbor multiple causal variants. However, current methods that address this problem assume that each locus contains a single causal variant. In this paper, we present eCAVIAR, a probabilistic method that has several key advantages over existing methods. First, our method can account for more than one causal variant in any given locus. Second, it can leverage summary statistics without accessing the individual genotype data. We use both simulated and real datasets to demonstrate the utility of our method. Using publicly available eQTL data on 45 different tissues, we demonstrate that eCAVIAR can prioritize likely relevant tissues and target genes for a set of glucose- and insulin-related trait loci.
Copyright © 2016 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


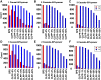
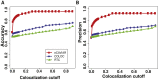
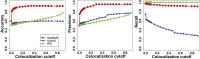
References
-
- Ripke S., O’Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M., Multicenter Genetic Studies of Schizophrenia Consortium. Psychosis Endophenotypes International Consortium. Wellcome Trust Case Control Consortium 2 Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01 ES021801/ES/NIEHS NIH HHS/United States
- R01 MH101782/MH/NIMH NIH HHS/United States
- K25 HL080079/HL/NHLBI NIH HHS/United States
- P30 NS062691/NS/NINDS NIH HHS/United States
- P01 HL028481/HL/NHLBI NIH HHS/United States
- U01 DA024417/DA/NIDA NIH HHS/United States
- U54 EB020403/EB/NIBIB NIH HHS/United States
- R01 ES022282/ES/NIEHS NIH HHS/United States
- R00 GM111744/GM/NIGMS NIH HHS/United States
- P01 HL030568/HL/NHLBI NIH HHS/United States
- HHSN268201000029C/HL/NHLBI NIH HHS/United States
- R01 GM083198/GM/NIGMS NIH HHS/United States

