Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity
- PMID: 27866838
- PMCID: PMC5742541
- DOI: 10.1016/j.cmet.2016.10.008
Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity
Abstract
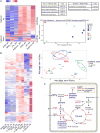
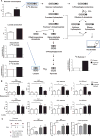
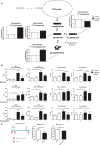
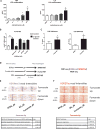
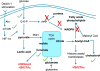
Induction of trained immunity (innate immune memory) is mediated by activation of immune and metabolic pathways that result in epigenetic rewiring of cellular functional programs. Through network-level integration of transcriptomics and metabolomics data, we identify glycolysis, glutaminolysis, and the cholesterol synthesis pathway as indispensable for the induction of trained immunity by β-glucan in monocytes. Accumulation of fumarate, due to glutamine replenishment of the TCA cycle, integrates immune and metabolic circuits to induce monocyte epigenetic reprogramming by inhibiting KDM5 histone demethylases. Furthermore, fumarate itself induced an epigenetic program similar to β-glucan-induced trained immunity. In line with this, inhibition of glutaminolysis and cholesterol synthesis in mice reduced the induction of trained immunity by β-glucan. Identification of the metabolic pathways leading to induction of trained immunity contributes to our understanding of innate immune memory and opens new therapeutic avenues.
Keywords: cholesterol metabolism; epigenetics; glutamine metabolism; glycolysis; trained immunity.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
References
-
- Chen Q, Giedt M, Tang L, Harrison DA. Tools and methods for studying the Drosophila JAK/STAT pathway. Methods. 2014;68:160–172. - PubMed
-
- Cheng SC, Scicluna BP, Arts RJ, Gresnigt MS, Lachmandas E, Giamarellos-Bourboulis EJ, Kox M, Manjeri GR, Wagenaars JA, Cremer OL, et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat Immunol. 2016;17:406–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

