Regulatory activities of transposable elements: from conflicts to benefits
- PMID: 27867194
- PMCID: PMC5498291
- DOI: 10.1038/nrg.2016.139
Regulatory activities of transposable elements: from conflicts to benefits
Abstract
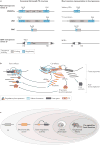
Transposable elements (TEs) are a prolific source of tightly regulated, biochemically active non-coding elements, such as transcription factor-binding sites and non-coding RNAs. Many recent studies reinvigorate the idea that these elements are pervasively co-opted for the regulation of host genes. We argue that the inherent genetic properties of TEs and the conflicting relationships with their hosts facilitate their recruitment for regulatory functions in diverse genomes. We review recent findings supporting the long-standing hypothesis that the waves of TE invasions endured by organisms for eons have catalysed the evolution of gene-regulatory networks. We also discuss the challenges of dissecting and interpreting the phenotypic effect of regulatory activities encoded by TEs in health and disease.
Figures


References
-
- McClintock B. Intranuclear systems controlling gene action and mutation. Brookhaven Symp Biol. 1956:58–74. - PubMed
-
- Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. - PubMed
-
- Britten RJ, Davidson EH. Repetitive and Non-Repetitive DNA Sequences and a Speculation on the Origins of Evolutionary Novelty. Q Rev Biol. 1971;46:111–138. - PubMed
-
- Orgel LE, Crick FH, Sapienza C. Selfish DNA. Nature. 1980;288:645–646. - PubMed
-
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

