NANOG reprograms prostate cancer cells to castration resistance via dynamically repressing and engaging the AR/FOXA1 signaling axis
- PMID: 27867534
- PMCID: PMC5109294
- DOI: 10.1038/celldisc.2016.41
NANOG reprograms prostate cancer cells to castration resistance via dynamically repressing and engaging the AR/FOXA1 signaling axis
Abstract
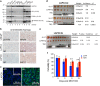
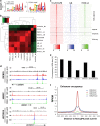
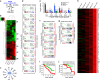
The pluripotency transcription factor NANOG has been implicated in tumor development, and NANOG-expressing cancer cells manifest stem cell properties that sustain tumor homeostasis, mediate therapy resistance and fuel tumor progression. However, how NANOG converges on somatic circuitry to trigger oncogenic reprogramming remains obscure. We previously reported that inducible NANOG expression propels the emergence of aggressive castration-resistant prostate cancer phenotypes. Here we first show that endogenous NANOG is required for the growth of castration-resistant prostate cancer xenografts. Genome-wide chromatin immunoprecipitation sequencing coupled with biochemical assays unexpectedly reveals that NANOG co-occupies a distinctive proportion of androgen receptor/Forkhead box A1 genomic loci and physically interacts with androgen receptor and Forkhead box A1. Integrative analysis of chromatin immunoprecipitation sequencing and time-resolved RNA sequencing demonstrates that NANOG dynamically alters androgen receptor/Forkhead box A1 signaling leading to both repression of androgen receptor-regulated pro-differentiation genes and induction of genes associated with cell cycle, stem cells, cell motility and castration resistance. Our studies reveal global molecular mechanisms whereby NANOG reprograms prostate cancer cells to a clinically relevant castration-resistant stem cell-like state driven by distinct NANOG-regulated gene clusters that correlate with patient survival. Thus, reprogramming factors such as NANOG may converge on and alter lineage-specific master transcription factors broadly in somatic cancers, thereby facilitating malignant disease progression and providing a novel route for therapeutic resistance.
Keywords: AR; FOXA1; NANOG; cancer stem cells; castration resistance; prostate cancer.
Figures




Similar articles
-
FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer.Am J Pathol. 2012 Feb;180(2):848-61. doi: 10.1016/j.ajpath.2011.10.021. Epub 2011 Dec 2. Am J Pathol. 2012. PMID: 22138582
-
Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer.Nucleic Acids Res. 2018 Feb 28;46(4):1895-1911. doi: 10.1093/nar/gkx1306. Nucleic Acids Res. 2018. PMID: 29309643 Free PMC article.
-
Transcriptional network of androgen receptor in prostate cancer progression.Int J Urol. 2013 Aug;20(8):756-68. doi: 10.1111/iju.12146. Epub 2013 Apr 21. Int J Urol. 2013. PMID: 23600948 Review.
-
Targeting Oct1 genomic function inhibits androgen receptor signaling and castration-resistant prostate cancer growth.Oncogene. 2016 Dec 8;35(49):6350-6358. doi: 10.1038/onc.2016.171. Epub 2016 Jun 6. Oncogene. 2016. PMID: 27270436
-
Role of prostate cancer stem-like cells in the development of antiandrogen resistance.Cancer Drug Resist. 2022 Jun 1;5(2):459-471. doi: 10.20517/cdr.2022.07. eCollection 2022. Cancer Drug Resist. 2022. PMID: 35800367 Free PMC article. Review.
Cited by
-
Cancer stem cells: Regulation programs, immunological properties and immunotherapy.Semin Cancer Biol. 2018 Oct;52(Pt 2):94-106. doi: 10.1016/j.semcancer.2018.05.001. Epub 2018 May 9. Semin Cancer Biol. 2018. PMID: 29752993 Free PMC article. Review.
-
LRIG1 is a pleiotropic androgen receptor-regulated feedback tumor suppressor in prostate cancer.Nat Commun. 2019 Dec 2;10(1):5494. doi: 10.1038/s41467-019-13532-4. Nat Commun. 2019. PMID: 31792211 Free PMC article.
-
The Multifaceted Role of Aldehyde Dehydrogenases in Prostate Cancer Stem Cells.Cancers (Basel). 2021 Sep 20;13(18):4703. doi: 10.3390/cancers13184703. Cancers (Basel). 2021. PMID: 34572930 Free PMC article. Review.
-
Comparative transcriptomics reveals a mixed basal, club, and hillock epithelial cell identity in castration-resistant prostate cancer.Proc Natl Acad Sci U S A. 2025 Feb 11;122(6):e2415308122. doi: 10.1073/pnas.2415308122. Epub 2025 Feb 6. Proc Natl Acad Sci U S A. 2025. PMID: 39913208 Free PMC article.
-
Cancer-associated fibroblasts and prostate cancer stem cells: crosstalk mechanisms and implications for disease progression.Front Cell Dev Biol. 2024 Jul 18;12:1412337. doi: 10.3389/fcell.2024.1412337. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39092186 Free PMC article. Review.
References
-
- Zhang J, Wang X, Li M et al. NANOGP8 is a retrogene expressed in cancers. FEBS J 2006; 273: 1723–1730. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

