Charge neutralization as the major factor for the assembly of nucleocapsid-like particles from C-terminal truncated hepatitis C virus core protein
- PMID: 27867765
- PMCID: PMC5111903
- DOI: 10.7717/peerj.2670
Charge neutralization as the major factor for the assembly of nucleocapsid-like particles from C-terminal truncated hepatitis C virus core protein
Abstract
Background: Hepatitis C virus (HCV) core protein, in addition to its structural role to form the nucleocapsid assembly, plays a critical role in HCV pathogenesis by interfering in several cellular processes, including microRNA and mRNA homeostasis. The C-terminal truncated HCV core protein (C124) is intrinsically unstructured in solution and is able to interact with unspecific nucleic acids, in the micromolar range, and to assemble into nucleocapsid-like particles (NLPs) in vitro. The specificity and propensity of C124 to the assembly and its implications on HCV pathogenesis are not well understood.
Methods: Spectroscopic techniques, transmission electron microscopy and calorimetry were used to better understand the propensity of C124 to fold or to multimerize into NLPs when subjected to different conditions or in the presence of unspecific nucleic acids of equivalent size to cellular microRNAs.
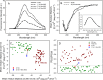
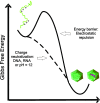
Results: The structural analysis indicated that C124 has low propensity to self-folding. On the other hand, for the first time, we show that C124, in the absence of nucleic acids, multimerizes into empty NLPs when subjected to a pH close to its isoelectric point (pH ≈ 12), indicating that assembly is mainly driven by charge neutralization. Isothermal calorimetry data showed that the assembly of NLPs promoted by nucleic acids is enthalpy driven. Additionally, data obtained from fluorescence correlation spectroscopy show that C124, in nanomolar range, was able to interact and to sequester a large number of short unspecific nucleic acids into NLPs.
Discussion: Together, our data showed that the charge neutralization is the major factor for the nucleocapsid-like particles assembly from C-terminal truncated HCV core protein. This finding suggests that HCV core protein may physically interact with unspecific cellular polyanions, which may correspond to microRNAs and mRNAs in a host cell infected by HCV, triggering their confinement into infectious particles.
Keywords: Capsid assembly; Circular dichroism; Fluorescence spectroscopy; HCV core protein; Structural biology.
Conflict of interest statement
Jerson Lima Silva is an Academic Editor for PeerJ.
Figures






Similar articles
-
Structural requirements for assembly and homotypic interactions of the hepatitis C virus core protein.Virus Res. 2006 Dec;122(1-2):137-43. doi: 10.1016/j.virusres.2006.07.008. Epub 2006 Sep 1. Virus Res. 2006. PMID: 16949699
-
The N-terminal half of the core protein of hepatitis C virus is sufficient for nucleocapsid formation.J Gen Virol. 2004 Apr;85(Pt 4):971-981. doi: 10.1099/vir.0.79775-0. J Gen Virol. 2004. PMID: 15039539
-
Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein.J Virol. 1990 Jul;64(7):3319-30. doi: 10.1128/JVI.64.7.3319-3330.1990. J Virol. 1990. PMID: 2191149 Free PMC article.
-
HCV core protein and virus assembly: what we know without structures.Immunol Res. 2014 Oct;60(1):1-10. doi: 10.1007/s12026-014-8494-3. Immunol Res. 2014. PMID: 24557493 Free PMC article. Review.
-
[Involvement of nonstructural protein 5A and lipids on production of hepatitis C virus particles].Uirusu. 2008 Dec;58(2):199-205. doi: 10.2222/jsv.58.199. Uirusu. 2008. PMID: 19374198 Review. Japanese.
Cited by
-
The interaction of dengue virus capsid protein with negatively charged interfaces drives the in vitro assembly of nucleocapsid-like particles.PLoS One. 2022 Mar 1;17(3):e0264643. doi: 10.1371/journal.pone.0264643. eCollection 2022. PLoS One. 2022. PMID: 35231063 Free PMC article.
-
Real-time compaction of nanoconfined DNA by an intrinsically disordered macromolecular counterion.Biochem Biophys Res Commun. 2020 Nov 26;533(1):175-180. doi: 10.1016/j.bbrc.2020.06.051. Epub 2020 Sep 18. Biochem Biophys Res Commun. 2020. PMID: 32951838 Free PMC article.
-
Elucidating the Thermodynamic Driving Forces of Polyanion-Templated Virus-like Particle Assembly.J Phys Chem B. 2019 Nov 21;123(46):9733-9741. doi: 10.1021/acs.jpcb.9b06258. Epub 2019 Nov 12. J Phys Chem B. 2019. PMID: 31661278 Free PMC article.
-
Self-assembly of dengue virus empty capsid-like particles in solution.iScience. 2023 Feb 14;26(3):106197. doi: 10.1016/j.isci.2023.106197. eCollection 2023 Mar 17. iScience. 2023. PMID: 36890794 Free PMC article.
References
-
- Acosta-Rivero N, Rodrigues A, Mussachio A, Poutu J, Falcon V, Torres D, Aguillar C, Linares M, Alonso M, Perez A, Menezes I, Morales-Grillo J, Marques G, Dueñas-Carrera S. A C-terminal truncated hepatitis C virus core protein variant assembles into virus-like particles in vitro in the absence of structured nucleic acids. Biochemical and Biophysical Research Communications. 2005;334:901–906. doi: 10.1016/j.bbrc.2005.06.185. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

