Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry
- PMID: 27869121
- PMCID: PMC5121339
- DOI: 10.1038/ncomms13404
Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry
Abstract
Although mutations may represent attractive targets for immunotherapy, direct identification of mutated peptide ligands isolated from human leucocyte antigens (HLA) on the surface of native tumour tissue has so far not been successful. Using advanced mass spectrometry (MS) analysis, we survey the melanoma-associated immunopeptidome to a depth of 95,500 patient-presented peptides. We thereby discover a large spectrum of attractive target antigen candidates including cancer testis antigens and phosphopeptides. Most importantly, we identify peptide ligands presented on native tumour tissue samples harbouring somatic mutations. Four of eleven mutated ligands prove to be immunogenic by neoantigen-specific T-cell responses. Moreover, tumour-reactive T cells with specificity for selected neoantigens identified by MS are detected in the patient's tumour and peripheral blood. We conclude that direct identification of mutated peptide ligands from primary tumour material by MS is possible and yields true neoepitopes with high relevance for immunotherapeutic strategies in cancer.
Figures
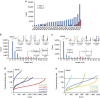

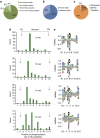



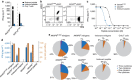
References
-
- Page D. B., Postow M. A., Callahan M. K., Allison J. P. & Wolchok J. D. Immune modulation in cancer with antibodies. Ann. Rev. Med. 65, 185–202 (2014). - PubMed
-
- Weber J. S. et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015). - PubMed
-
- van Baren N. et al. Tumoral and immunologic response after vaccination of melanoma patients with an ALVAC virus encoding MAGE antigens recognized by T cells. J. Clin. Oncol.: Off. J. Am. Soc. Clin. Oncol. 23, 9008–9021 (2005). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

