Reprogramming mouse fibroblasts into engraftable myeloerythroid and lymphoid progenitors
- PMID: 27869129
- PMCID: PMC5121332
- DOI: 10.1038/ncomms13396
Reprogramming mouse fibroblasts into engraftable myeloerythroid and lymphoid progenitors
Abstract
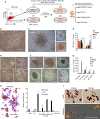
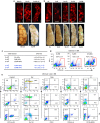
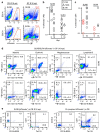
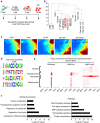
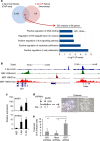
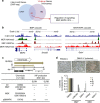
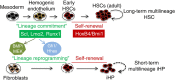
Recent efforts have attempted to convert non-blood cells into hematopoietic stem cells (HSCs) with the goal of generating blood lineages de novo. Here we show that hematopoietic transcription factors Scl, Lmo2, Runx1 and Bmi1 can convert a developmentally distant lineage (fibroblasts) into 'induced hematopoietic progenitors' (iHPs). Functionally, iHPs generate acetylcholinesterase+ megakaryocytes and phagocytic myeloid cells in vitro and can also engraft immunodeficient mice, generating myeloerythoid and B-lymphoid cells for up to 4 months in vivo. Molecularly, iHPs transcriptionally resemble native Kit+ hematopoietic progenitors. Mechanistically, reprogramming factor Lmo2 implements a hematopoietic programme in fibroblasts by rapidly binding to and upregulating the Hhex and Gfi1 genes within days. Moreover the reprogramming transcription factors also require extracellular BMP and MEK signalling to cooperatively effectuate reprogramming. Thus, the transcription factors that orchestrate embryonic hematopoiesis can artificially reconstitute this programme in developmentally distant fibroblasts, converting them into engraftable blood progenitors.
Figures

References
-
- Graf T. & Enver T. Forcing cells to change lineages. Nature 462, 587–594 (2009). - PubMed
-
- Davis R. L., Weintraub H. & Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). - PubMed
-
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

