Synthetic recording and in situ readout of lineage information in single cells
- PMID: 27869821
- PMCID: PMC6487260
- DOI: 10.1038/nature20777
Synthetic recording and in situ readout of lineage information in single cells
Abstract
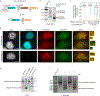
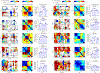
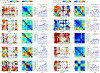
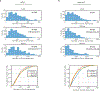
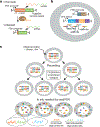
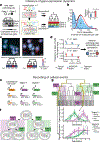
Reconstructing the lineage relationships and dynamic event histories of individual cells within their native spatial context is a long-standing challenge in biology. Many biological processes of interest occur in optically opaque or physically inaccessible contexts, necessitating approaches other than direct imaging. Here we describe a synthetic system that enables cells to record lineage information and event histories in the genome in a format that can be subsequently read out of single cells in situ. This system, termed memory by engineered mutagenesis with optical in situ readout (MEMOIR), is based on a set of barcoded recording elements termed scratchpads. The state of a given scratchpad can be irreversibly altered by CRISPR/Cas9-based targeted mutagenesis, and later read out in single cells through multiplexed single-molecule RNA fluorescence hybridization (smFISH). Using MEMOIR as a proof of principle, we engineered mouse embryonic stem cells to contain multiple scratchpads and other recording components. In these cells, scratchpads were altered in a progressive and stochastic fashion as the cells proliferated. Analysis of the final states of scratchpads in single cells in situ enabled reconstruction of lineage information from cell colonies. Combining analysis of endogenous gene expression with lineage reconstruction in the same cells further allowed inference of the dynamic rates at which embryonic stem cells switch between two gene expression states. Finally, using simulations, we show how parallel MEMOIR systems operating in the same cell could enable recording and readout of dynamic cellular event histories. MEMOIR thus provides a versatile platform for information recording and in situ, single-cell readout across diverse biological systems.
Conflict of interest statement
The authors declare competing financial interests: details are available in the online version of the paper.
Figures








Comment in
-
Systems biology: Molecular memoirs of a cellular family.Nature. 2017 Jan 5;541(7635):38-39. doi: 10.1038/nature21101. Epub 2016 Dec 14. Nature. 2017. PMID: 27974796 No abstract available.

