Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias
- PMID: 27869828
- PMCID: PMC5206905
- DOI: 10.1038/ng.3726
Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias
Abstract
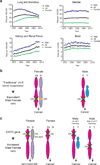
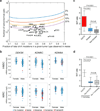
There is a striking and unexplained male predominance across many cancer types. A subset of X-chromosome genes can escape X-inactivation, which would protect females from complete functional loss by a single mutation. To identify putative 'escape from X-inactivation tumor-suppressor' (EXITS) genes, we examined somatic alterations from >4,100 cancers across 21 tumor types for sex bias. Six of 783 non-pseudoautosomal region (PAR) X-chromosome genes (ATRX, CNKSR2, DDX3X, KDM5C, KDM6A, and MAGEC3) harbored loss-of-function mutations more frequently in males (based on a false discovery rate < 0.1), in comparison to zero of 18,055 autosomal and PAR genes (Fisher's exact P < 0.0001). Male-biased mutations in genes that escape X-inactivation were observed in combined analysis across many cancers and in several individual tumor types, suggesting a generalized phenomenon. We conclude that biallelic expression of EXITS genes in females explains a portion of the reduced cancer incidence in females as compared to males across a variety of tumor types.
Figures

Comment in
-
An EXITS strategy for decreasing cancer risk in women.Sci Transl Med. 2016 Dec 7;8(368):368ec197. doi: 10.1126/scitranslmed.aal2806. Sci Transl Med. 2016. PMID: 27928023 No abstract available.
-
Cancer genetics: X-inactivation and cancer incidence.Nat Rev Cancer. 2016 Dec 21;17(1):3. doi: 10.1038/nrc.2016.152. Nat Rev Cancer. 2016. PMID: 27999427 No abstract available.
References
-
- Edgren G, Liang L, Adami HO, Chang ET. Enigmatic sex disparities in cancer incidence. European journal of epidemiology. 2012;27:187–196. - PubMed
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11–30. - PubMed
-
- Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

