Antizyme inhibitor 1: a potential carcinogenic molecule
- PMID: 27870265
- PMCID: PMC5329145
- DOI: 10.1111/cas.13122
Antizyme inhibitor 1: a potential carcinogenic molecule
Abstract
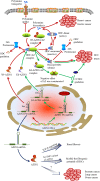
Polyamines are multivalent and organic cations essential for cellular growth, proliferation, differentiation, and apoptosis. Increased levels of polyamines are closely associated with numerous forms of cancer. An autoregulatory circuit composed of ornithine decarboxylase (ODC), antizyme (AZ) and antizyme inhibitor (AZI) govern the intracellular level of polyamines. Antizyme binds with ODC to inhibit ODC activity and to promote the ubiquitin-independent degradation of ODC. Antizyme inhibitor binds to AZ with a higher affinity than ODC. Consequently, ODC is released from the ODC-AZ complex to rescue its activity. Antizyme inhibitor increases the ODC activity to accelerate the formation of intracellular polyamines, triggering gastric and breast carcinogenesis as well as hepatocellular carcinoma and esophageal squamous cell carcinoma development. Antizyme inhibitor 1 (AZIN1), a primary member of the AZI family, has aroused more attention because of its contribution to cancer. Even though its conformation is changed by adenosine-to-inosine (A→I) RNA editing, it plays an important role in tumorigenesis through regulating intracellular polyamines. Encouragingly, AZIN1 has been revealed to have an additional function outside the polyamine pathway so as to bypass the deficiency of targeting the polyamine biosynthetic pathway, promising to become a critical target for cancer therapy. Here, we review the latest research advances into AZIN1 and its potential contribution to carcinogenesis.
Keywords: Antizyme; antizyme inhibitor 1; carcinogenesis; ornithine decarboxylase; polyamine.
© 2016 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures
Similar articles
-
Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor.Essays Biochem. 2009 Nov 4;46:47-61. doi: 10.1042/bse0460004. Essays Biochem. 2009. PMID: 20095969 Review.
-
The antizyme family for regulating polyamines.J Biol Chem. 2018 Nov 30;293(48):18730-18735. doi: 10.1074/jbc.TM118.003339. Epub 2018 Oct 24. J Biol Chem. 2018. PMID: 30355739 Free PMC article. Review.
-
Stable siRNA-mediated silencing of antizyme inhibitor: regulation of ornithine decarboxylase activity.Biochem Biophys Res Commun. 2005 Mar 4;328(1):206-12. doi: 10.1016/j.bbrc.2004.11.172. Biochem Biophys Res Commun. 2005. PMID: 15670771
-
Protein degradation, the main hub in the regulation of cellular polyamines.Biochem J. 2016 Dec 15;473(24):4551-4558. doi: 10.1042/BCJ20160519C. Biochem J. 2016. PMID: 27941031 Review.
-
Identification, assay, and functional analysis of the antizyme inhibitor family.Methods Mol Biol. 2011;720:269-78. doi: 10.1007/978-1-61779-034-8_16. Methods Mol Biol. 2011. PMID: 21318879
Cited by
-
Antizyme inhibitor 1 genetic polymorphisms associated with diabetic patients validated in the livers of diabetic mice.Exp Ther Med. 2019 Oct;18(4):3139-3146. doi: 10.3892/etm.2019.7919. Epub 2019 Aug 20. Exp Ther Med. 2019. PMID: 31572554 Free PMC article.
-
Ornithine decarboxylase antizyme inhibitor 2 (AZIN2) is a signature of secretory phenotype and independent predictor of adverse prognosis in colorectal cancer.PLoS One. 2019 Feb 15;14(2):e0211564. doi: 10.1371/journal.pone.0211564. eCollection 2019. PLoS One. 2019. PMID: 30768610 Free PMC article.
-
Polyamines: Functions, Metabolism, and Role in Human Disease Management.Med Sci (Basel). 2021 Jun 9;9(2):44. doi: 10.3390/medsci9020044. Med Sci (Basel). 2021. PMID: 34207607 Free PMC article. Review.
-
EPLIN-β is a novel substrate of ornithine decarboxylase antizyme 1 and mediates cellular migration.J Cell Sci. 2023 Jun 15;136(12):jcs260427. doi: 10.1242/jcs.260427. Epub 2023 Jun 16. J Cell Sci. 2023. PMID: 37325974 Free PMC article.
-
Unveiling the hidden players: noncoding RNAs orchestrating polyamine metabolism in disease.Cell Biosci. 2024 Jun 25;14(1):84. doi: 10.1186/s13578-024-01235-3. Cell Biosci. 2024. PMID: 38918813 Free PMC article. Review.
References
-
- Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem 2006; 281: 14529–32. - PubMed
-
- Kahana C, Asher G, Shaul Y. Mechanisms of protein degradation: an odyssey with ODC. Cell Cycle 2005; 4: 1461–4. - PubMed
-
- Gerner EW, Meyskens FL. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer 2004; 4: 781–92. - PubMed
-
- Snapir Z, Keren‐Paz A, Bercovich Z, Kahana C. Antizyme 3 inhibits polyamine uptake and ornithine decarboxylase (ODC) activity, but does not stimulate ODC degradation. Biochem J 2009; 419: 99–103, 101 p following 103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

