Position-dependent termination and widespread obligatory frameshifting in Euplotes translation
- PMID: 27870834
- PMCID: PMC5295771
- DOI: 10.1038/nsmb.3330
Position-dependent termination and widespread obligatory frameshifting in Euplotes translation
Abstract
The ribosome can change its reading frame during translation in a process known as programmed ribosomal frameshifting. These rare events are supported by complex mRNA signals. However, we found that the ciliates Euplotes crassus and Euplotes focardii exhibit widespread frameshifting at stop codons. 47 different codons preceding stop signals resulted in either +1 or +2 frameshifts, and +1 frameshifting at AAA was the most frequent. The frameshifts showed unusual plasticity and rapid evolution, and had little influence on translation rates. The proximity of a stop codon to the 3' mRNA end, rather than its occurrence or sequence context, appeared to designate termination. Thus, a 'stop codon' is not a sufficient signal for translation termination, and the default function of stop codons in Euplotes is frameshifting, whereas termination is specific to certain mRNA positions and probably requires additional factors.
Figures
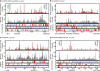


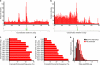
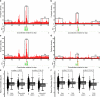

References
-
- Baranov PV, Atkins JF, Yordanova MM. Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nature Rev Genetics. 2015;16:517–529. - PubMed
-
- Klobutcher LA, Farabaugh PJ. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell. 2002;111:763–766. - PubMed
Methods-only References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

