Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis
- PMID: 27871231
- PMCID: PMC5117596
- DOI: 10.1186/s12864-016-3293-y
Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis
Abstract
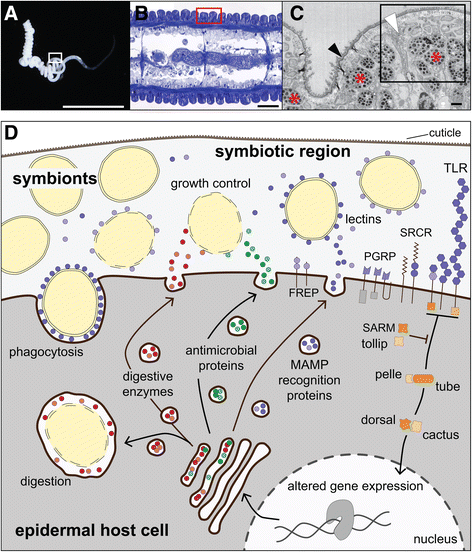
Background: The gutless marine worm Olavius algarvensis has a completely reduced digestive and excretory system, and lives in an obligate nutritional symbiosis with bacterial symbionts. While considerable knowledge has been gained of the symbionts, the host has remained largely unstudied. Here, we generated transcriptomes and proteomes of O. algarvensis to better understand how this annelid worm gains nutrition from its symbionts, how it adapted physiologically to a symbiotic lifestyle, and how its innate immune system recognizes and responds to its symbiotic microbiota.
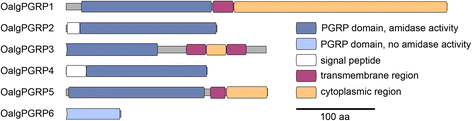
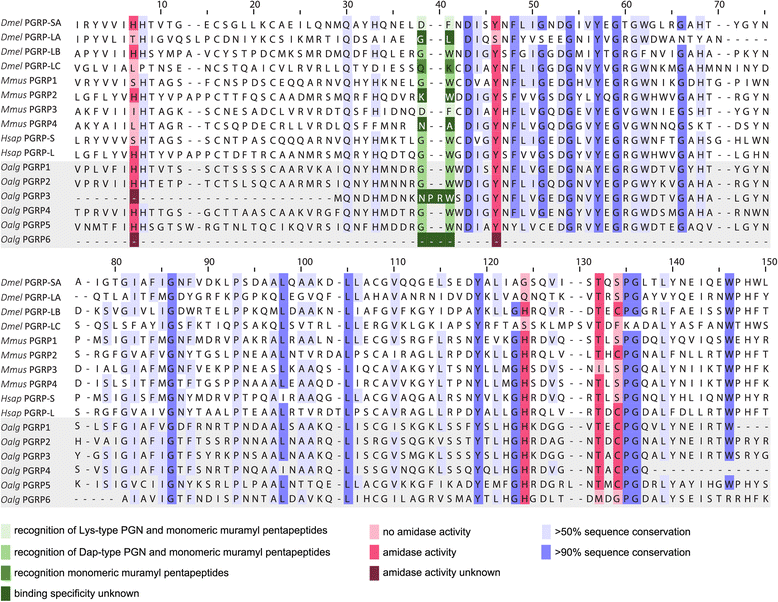
Results: Key adaptations to the symbiosis include (i) the expression of gut-specific digestive enzymes despite the absence of a gut, most likely for the digestion of symbionts in the host's epidermal cells; (ii) a modified hemoglobin that may bind hydrogen sulfide produced by two of the worm's symbionts; and (iii) the expression of a very abundant protein for oxygen storage, hemerythrin, that could provide oxygen to the symbionts and the host under anoxic conditions. Additionally, we identified a large repertoire of proteins involved in interactions between the worm's innate immune system and its symbiotic microbiota, such as peptidoglycan recognition proteins, lectins, fibrinogen-related proteins, Toll and scavenger receptors, and antimicrobial proteins.
Conclusions: We show how this worm, over the course of evolutionary time, has modified widely-used proteins and changed their expression patterns in adaptation to its symbiotic lifestyle and describe expressed components of the innate immune system in a marine oligochaete. Our results provide further support for the recent realization that animals have evolved within the context of their associations with microbes and that their adaptive responses to symbiotic microbiota have led to biological innovations.
Keywords: Annelida; Carbon monoxide; Chemosynthetic symbiosis; FREP; Immunology; Oligochaeta; PGRP; Phallodrilinae; RNA-Seq; Respiratory pigment; SRCR.
Figures

Similar articles
-
Symbiosis insights through metagenomic analysis of a microbial consortium.Nature. 2006 Oct 26;443(7114):950-5. doi: 10.1038/nature05192. Epub 2006 Sep 17. Nature. 2006. PMID: 16980956
-
Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use.Proc Natl Acad Sci U S A. 2012 May 8;109(19):E1173-82. doi: 10.1073/pnas.1121198109. Epub 2012 Apr 18. Proc Natl Acad Sci U S A. 2012. PMID: 22517752 Free PMC article.
-
Coexistence of bacterial sulfide oxidizers, sulfate reducers, and spirochetes in a gutless worm (Oligochaeta) from the Peru margin.Appl Environ Microbiol. 2005 Mar;71(3):1553-61. doi: 10.1128/AEM.71.3.1553-1561.2005. Appl Environ Microbiol. 2005. PMID: 15746360 Free PMC article.
-
Role of antimicrobial peptides in controlling symbiotic bacterial populations.Nat Prod Rep. 2018 Apr 25;35(4):336-356. doi: 10.1039/c7np00056a. Nat Prod Rep. 2018. PMID: 29393944 Review.
-
Understanding regulation of the host-mediated gut symbiont population and the symbiont-mediated host immunity in the Riptortus-Burkholderia symbiosis system.Dev Comp Immunol. 2016 Nov;64:75-81. doi: 10.1016/j.dci.2016.01.005. Epub 2016 Jan 13. Dev Comp Immunol. 2016. PMID: 26774501 Review.
Cited by
-
Fidelity varies in the symbiosis between a gutless marine worm and its microbial consortium.Microbiome. 2022 Oct 22;10(1):178. doi: 10.1186/s40168-022-01372-2. Microbiome. 2022. PMID: 36273146 Free PMC article.
-
Metabarcoding reveals distinct microbiotypes in the giant clam Tridacna maxima.Microbiome. 2020 Apr 21;8(1):57. doi: 10.1186/s40168-020-00835-8. Microbiome. 2020. PMID: 32317019 Free PMC article.
-
Genomic Signatures Supporting the Symbiosis and Formation of Chitinous Tube in the Deep-Sea Tubeworm Paraescarpia echinospica.Mol Biol Evol. 2021 Sep 27;38(10):4116-4134. doi: 10.1093/molbev/msab203. Mol Biol Evol. 2021. PMID: 34255082 Free PMC article.
-
Distinct genomic routes underlie transitions to specialised symbiotic lifestyles in deep-sea annelid worms.Nat Commun. 2023 May 17;14(1):2814. doi: 10.1038/s41467-023-38521-6. Nat Commun. 2023. PMID: 37198188 Free PMC article.
-
Considerations for constructing a protein sequence database for metaproteomics.Comput Struct Biotechnol J. 2022 Jan 21;20:937-952. doi: 10.1016/j.csbj.2022.01.018. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35242286 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

