Transcriptome analysis reveals manifold mechanisms of cyst development in ADPKD
- PMID: 27871310
- PMCID: PMC5117508
- DOI: 10.1186/s40246-016-0095-x
Transcriptome analysis reveals manifold mechanisms of cyst development in ADPKD
Abstract
Background: Autosomal dominant polycystic kidney disease (ADPKD) causes progressive loss of renal function in adults as a consequence of the accumulation of cysts. ADPKD is the most common genetic cause of end-stage renal disease. Mutations in polycystin-1 occur in 87% of cases of ADPKD and mutations in polycystin-2 are found in 12% of ADPKD patients. The complexity of ADPKD has hampered efforts to identify the mechanisms underlying its pathogenesis. No current FDA (Federal Drug Administration)-approved therapies ameliorate ADPKD progression.
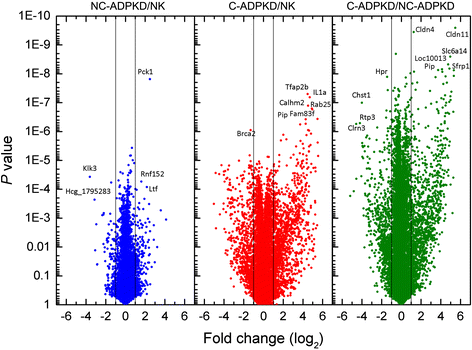
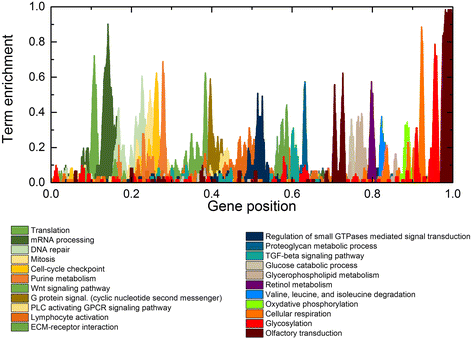
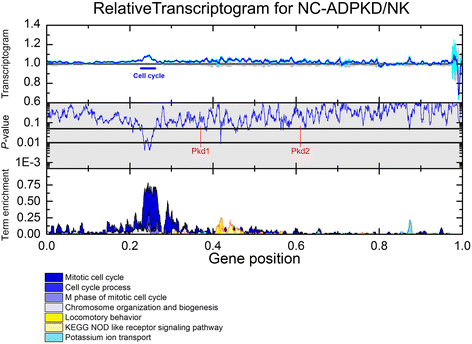
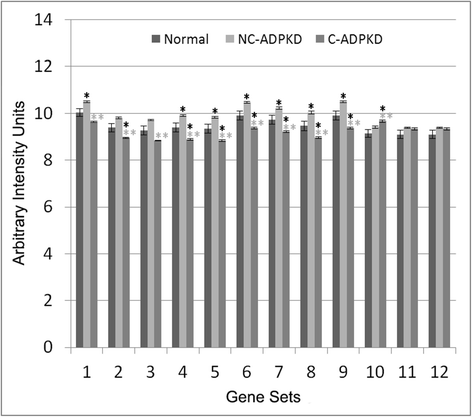
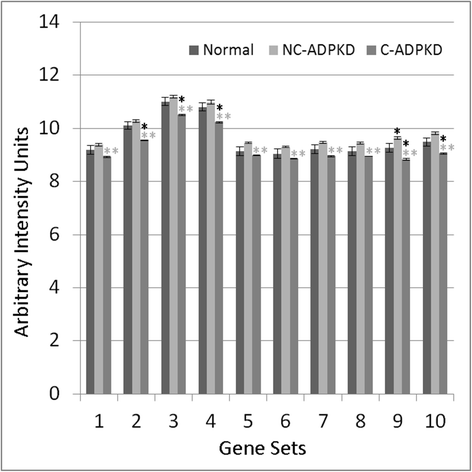
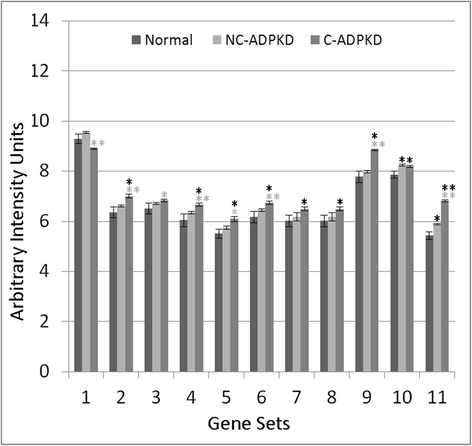
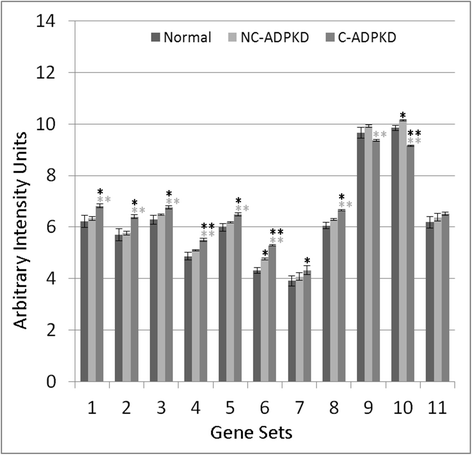
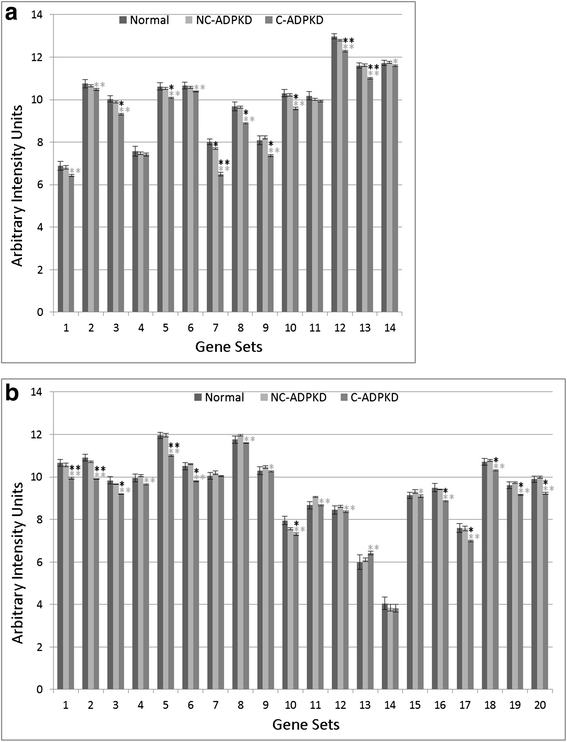
Results: We used the de Almeida laboratory's sensitive new transcriptogram method for whole-genome gene expression data analysis to analyze microarray data from cell lines developed from cell isolates of normal kidney and of both non-cystic nephrons and cysts from the kidney of a patient with ADPKD. We compared results obtained using standard Ingenuity Volcano plot analysis, Gene Set Enrichment Analysis (GSEA) and transcriptogram analysis. Transcriptogram analysis confirmed the findings of Ingenuity, GSEA, and published analysis of ADPKD kidney data and also identified multiple new expression changes in KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways related to cell growth, cell death, genetic information processing, nucleotide metabolism, signal transduction, immune response, response to stimulus, cellular processes, ion homeostasis and transport and cofactors, vitamins, amino acids, energy, carbohydrates, drugs, lipids, and glycans. Transcriptogram analysis also provides significance metrics which allow us to prioritize further study of these pathways.
Conclusions: Transcriptogram analysis identifies novel pathways altered in ADPKD, providing new avenues to identify both ADPKD's mechanisms of pathogenesis and pharmaceutical targets to ameliorate the progression of the disease.
Keywords: Autosomal dominant polycystic kidney disease; Bioinformatics; Cystic kidney disease; Kidney; Pathway identification; Transcriptogram.
Figures







Similar articles
-
Parallel microarray profiling identifies ErbB4 as a determinant of cyst growth in ADPKD and a prognostic biomarker for disease progression.Am J Physiol Renal Physiol. 2017 Apr 1;312(4):F577-F588. doi: 10.1152/ajprenal.00607.2016. Epub 2017 Jan 11. Am J Physiol Renal Physiol. 2017. PMID: 28077374 Free PMC article.
-
Polycystin-2-dependent transcriptome reveals early response of autosomal dominant polycystic kidney disease.Physiol Genomics. 2023 Nov 1;55(11):565-577. doi: 10.1152/physiolgenomics.00040.2023. Epub 2023 Sep 18. Physiol Genomics. 2023. PMID: 37720991 Free PMC article.
-
Profiling conserved biological pathways in Autosomal Dominant Polycystic Kidney Disorder (ADPKD) to elucidate key transcriptomic alterations regulating cystogenesis: A cross-species meta-analysis approach.Gene. 2017 Sep 5;627:434-450. doi: 10.1016/j.gene.2017.06.059. Epub 2017 Jul 1. Gene. 2017. PMID: 28676447
-
Molecular pathogenesis of autosomal dominant polycystic kidney disease.Expert Rev Mol Med. 2006 Jan 17;8(2):1-22. doi: 10.1017/S1462399406010362. Expert Rev Mol Med. 2006. PMID: 16515728 Review.
-
Cell Proliferation and Apoptosis in ADPKD.Adv Exp Med Biol. 2016;933:25-34. doi: 10.1007/978-981-10-2041-4_3. Adv Exp Med Biol. 2016. PMID: 27730432 Review.
Cited by
-
Super-enhancer-driven metabolic reprogramming promotes cystogenesis in autosomal dominant polycystic kidney disease.Nat Metab. 2020 Aug;2(8):717-731. doi: 10.1038/s42255-020-0227-4. Epub 2020 Jul 13. Nat Metab. 2020. PMID: 32694829
-
Predicting Progression of Autosomal Dominant Polycystic Kidney Disease by Changes in the Telomeric Epigenome.Cells. 2022 Oct 20;11(20):3300. doi: 10.3390/cells11203300. Cells. 2022. PMID: 36291168 Free PMC article.
-
Transcriptogram analysis reveals relationship between viral titer and gene sets responses during Corona-virus infection.NAR Genom Bioinform. 2022 Mar 15;4(1):lqac020. doi: 10.1093/nargab/lqac020. eCollection 2022 Mar. NAR Genom Bioinform. 2022. PMID: 35300459 Free PMC article.
-
Role of chemokines, innate and adaptive immunity.Cell Signal. 2020 Sep;73:109647. doi: 10.1016/j.cellsig.2020.109647. Epub 2020 Apr 20. Cell Signal. 2020. PMID: 32325183 Free PMC article. Review.
-
Clinical, histopathologic and molecular features of idiopathic and diabetic nodular mesangial sclerosis in humans.Nephrol Dial Transplant. 2021 Dec 31;37(1):72-84. doi: 10.1093/ndt/gfaa331. Nephrol Dial Transplant. 2021. PMID: 33537765 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

