Conformational Control of Cascade Interference and Priming Activities in CRISPR Immunity
- PMID: 27871367
- PMCID: PMC5561731
- DOI: 10.1016/j.molcel.2016.09.033
Conformational Control of Cascade Interference and Priming Activities in CRISPR Immunity
Abstract
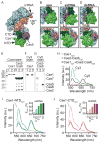
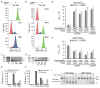
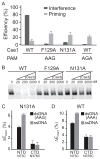
During type I-E CRISPR-Cas immunity, the Cascade surveillance complex utilizes CRISPR-derived RNAs to target complementary invasive DNA for destruction. When invader mutation blocks this interference activity, Cascade instead triggers rapid primed adaptation against the invader. The molecular basis for this dual Cascade activity is poorly understood. Here we show that the conformation of the Cse1 subunit controls Cascade activity. Using FRET, we find that Cse1 exists in a dynamic equilibrium between "open" and "closed" conformations, and the extent to which the open conformation is favored directly correlates with the attenuation of interference and relative increase in priming activity upon target mutation. Additionally, the Cse1 L1 motif modulates Cascade activity by stabilizing the closed conformation. L1 mutations promote the open conformation and switch immune response from interference to priming. Our results demonstrate that Cascade conformation controls the functional outcome of target recognition, enabling tunable CRISPR immune response to combat invader evolution.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
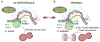
Comment in
-
Memory Upgrade: Insights into Primed Adaptation by CRISPR-Cas Immune Systems.Mol Cell. 2016 Nov 17;64(4):641-642. doi: 10.1016/j.molcel.2016.11.008. Mol Cell. 2016. PMID: 27863223 Free PMC article.
References
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science (80– ) 2007;315:1709–1712. - PubMed
-
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

