Diversity of bat astroviruses in Lao PDR and Cambodia
- PMID: 27871796
- PMCID: PMC7106329
- DOI: 10.1016/j.meegid.2016.11.013
Diversity of bat astroviruses in Lao PDR and Cambodia
Abstract
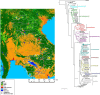
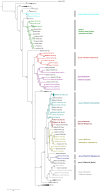
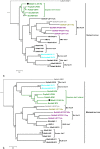
Astroviruses are known to infect humans and a wide range of animal species, and can cause gastroenteritis in their hosts. Recent studies have reported astroviruses in bats in Europe and in several locations in China. We sampled 1876 bats from 17 genera at 45 sites from 14 and 13 provinces in Cambodia and Lao PDR respectively, and tested them for astroviruses. Our study revealed a high diversity of astroviruses among various Yangochiroptera and Yinpterochiroptera bats. Evidence for varying degrees of host restriction for astroviruses in bats was found. Furthermore, additional Pteropodid hosts were detected. The astroviruses formed distinct phylogenetic clusters within the genus Mamastrovirus, most closely related to other known bat astroviruses. The astrovirus sequences were found to be highly saturated indicating that phylogenetic relationships should be interpreted carefully. An astrovirus clustering in a group with other viruses from diverse hosts, including from ungulates and porcupines, was found in a Rousettus bat. These findings suggest that diverse astroviruses can be found in many species of mammals, including bats.
Keywords: Astroviruses; Bats; Cambodia; Genetic diversity; Lao PDR.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures
References
-
- Bates P., Bumrungsri S., Francis C., Csorba G. Hipposideros armiger [WWW Document]. IUCN red list threat. Species 2008 ET10110A3162617. 2008. (URL, accessed 9.21.16) - DOI
-
- Bates P., Helgen K. Rousettus leschenaultii [WWW Document]. IUCN red list threat. Species 2008 ET19756A9011055. 2008. (URL, accessed 11.4.16) - DOI
-
- Bates P., Kingston T., Francis C., Rosell-Ambal G., Heaney L., Gonzales J.C., Molur S., Srinivasulu C. Scotophilus kuhlii [WWW Document]. IUCN Red List Threat. Species 2008 ET20068A9142479. 2008. (URL, accessed 9.21.16) - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

