Genome-wide minor histocompatibility matching as related to the risk of graft-versus-host disease
- PMID: 27872059
- PMCID: PMC5301826
- DOI: 10.1182/blood-2016-09-737700
Genome-wide minor histocompatibility matching as related to the risk of graft-versus-host disease
Abstract
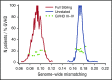
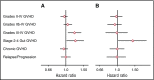
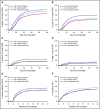
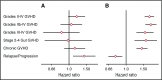
The risk of acute graft-versus-host disease (GVHD) is higher after allogeneic hematopoietic cell transplantation (HCT) from unrelated donors as compared with related donors. This difference has been explained by increased recipient mismatching for major histocompatibility antigens or minor histocompatibility antigens. In the current study, we used genome-wide arrays to enumerate single nucleotide polymorphisms (SNPs) that produce graft-versus-host (GVH) amino acid coding differences between recipients and donors. We then tested the hypothesis that higher degrees of genome-wide recipient GVH mismatching correlate with higher risks of GVHD after allogeneic HCT. In HLA-genotypically matched sibling recipients, the average recipient mismatching of coding SNPs was 9.35%. Each 1% increase in genome-wide recipient mismatching was associated with an estimated 20% increase in the hazard of grades III-IV GVHD (hazard ratio [HR], 1.20; 95% confidence interval [CI], 1.05-1.37; P = .007) and an estimated 22% increase in the hazard of stage 2-4 acute gut GVHD (HR, 1.22; 95% CI, 1.02-1.45; P = .03). In HLA-A, B, C, DRB1, DQA1, DQB1, DPA1, DPB1-phenotypically matched unrelated recipients, the average recipient mismatching of coding SNPs was 17.3%. The estimated risks of GVHD-related outcomes in HLA-phenotypically matched unrelated recipients were low, relative to the large difference in genome-wide mismatching between the 2 groups. In contrast, the risks of GVHD-related outcomes were higher in HLA-DP GVH-mismatched unrelated recipients than in HLA-matched sibling recipients. Taken together, these results suggest that the increased GVHD risk after unrelated HCT is predominantly an effect of HLA-mismatching.
© 2017 by The American Society of Hematology.
Figures

Comment in
-
Major vs minor histocompatibility antigens.Blood. 2017 Feb 9;129(6):664-666. doi: 10.1182/blood-2016-12-754515. Blood. 2017. PMID: 28183746 No abstract available.
References
-
- Weir BS, Anderson AD, Hepler AB. Genetic relatedness analysis: modern data and new challenges. Nat Rev Genet. 2006;7(10):771-780. - PubMed
-
- Martin PJ. Increased disparity for minor histocompatibility antigens as a potential cause of increased GVHD risk in marrow transplantation from unrelated donors compared with related donors. Bone Marrow Transplant. 1991;8(3):217-223. - PubMed
-
- Mullally A, Ritz J. Beyond HLA: the significance of genomic variation for allogeneic hematopoietic stem cell transplantation. Blood. 2007;109(4):1355-1362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

