Cytotoxic 1-deoxysphingolipids are metabolized by a cytochrome P450-dependent pathway
- PMID: 27872144
- PMCID: PMC5234722
- DOI: 10.1194/jlr.M072421
Cytotoxic 1-deoxysphingolipids are metabolized by a cytochrome P450-dependent pathway
Abstract
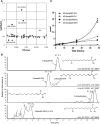
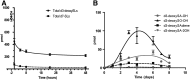
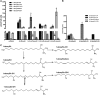
The 1-deoxysphingolipids (1-deoxySLs) are atypical sphingolipids (SLs) that are formed when serine palmitoyltransferase condenses palmitoyl-CoA with alanine instead of serine during SL synthesis. The 1-deoxySLs are toxic to neurons and pancreatic β-cells. Pathologically elevated 1-deoxySLs cause the inherited neuropathy, hereditary sensory autonomic neuropathy type 1 (HSAN1), and are also found in T2D. Diabetic sensory polyneuropathy (DSN) and HSAN1 are clinically very similar, suggesting that 1-deoxySLs may be implicated in both pathologies. The 1-deoxySLs are considered to be dead-end metabolites, as they lack the C1-hydroxyl group, which is essential for the canonical degradation of SLs. Here, we report a previously unknown metabolic pathway, which is capable of degrading 1-deoxySLs. Using a variety of metabolic labeling approaches and high-resolution high-accuracy MS, we identified eight 1-deoxySL downstream metabolites, which appear to be formed by cytochrome P450 (CYP)4F enzymes. Comprehensive inhibition and induction of CYP4F enzymes blocked and stimulated, respectively, the formation of the downstream metabolites. Consequently, CYP4F enzymes might be novel therapeutic targets for the treatment of HSAN1 and DSN, as well as for the prevention of T2D.
Keywords: diabetes; lipids/chemistry; mass spectrometry; metabolic pathway; neurotoxicity; obesity; sphingolipids.
Copyright © 2017 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures

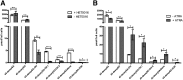

References
-
- Weiss B., and Stoffel W.. 1997. Human and murine serine-palmitoyl-CoA transferase–cloning, expression and characterization of the key enzyme in sphingolipid synthesis. Eur. J. Biochem. 249: 239–247. - PubMed
-
- Hanada K., Hara T., Nishijima M., Kuge O., Dickson R. C., and Nagiec M. M.. 1997. A mammalian homolog of the yeast LCB1 encodes a component of serine palmitoyltransferase, the enzyme catalyzing the first step in sphingolipid synthesis. J. Biol. Chem. 272: 32108–32114. - PubMed
-
- Zitomer N. C., Mitchell T., Voss K. A., Bondy G. S., Pruett S. T., Garnier-Amblard E. C., Liebeskind L. S., Park H., Wang E., Sullards M. C., et al. . 2009. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: a novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J. Biol. Chem. 284: 4786–4795. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

