Biophysical Evidence for Intrinsic Disorder in the C-terminal Tails of the Epidermal Growth Factor Receptor (EGFR) and HER3 Receptor Tyrosine Kinases
- PMID: 27872189
- PMCID: PMC5241735
- DOI: 10.1074/jbc.M116.747485
Biophysical Evidence for Intrinsic Disorder in the C-terminal Tails of the Epidermal Growth Factor Receptor (EGFR) and HER3 Receptor Tyrosine Kinases
Abstract
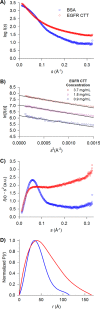
The epidermal growth factor receptor (EGFR)/ErbB family of receptor tyrosine kinases includes oncogenes important in the progression of breast and other cancers, and they are targets for many drug development strategies. Each member of the ErbB family possesses a unique, structurally uncharacterized C-terminal tail that plays an important role in autophosphorylation and signal propagation. To determine whether these C-terminal tails are intrinsically disordered regions, we conducted a battery of biophysical experiments on the EGFR and HER3 tails. Using hydrogen/deuterium exchange mass spectrometry, we measured the conformational dynamics of intracellular half constructs and compared the tails with the ordered kinase domains. The C-terminal tails demonstrate more rapid deuterium exchange behavior when compared with the kinase domains. Next, we expressed and purified EGFR and HER3 tail-only constructs. Results from circular dichroism spectroscopy, size exclusion chromatography with multiangle light scattering, dynamic light scattering, analytical ultracentrifugation, and small angle X-ray scattering each provide evidence that the EGFR and HER3 C-terminal tails are intrinsically disordered with extended, non-globular structure in solution. The intrinsic disorder and extended conformation of these tails may be important for their function by increasing the capture radius and reducing the thermodynamic barriers for binding of downstream signaling proteins.
Keywords: C-terminal domain (carboxyl tail domain, CTD); analytical ultracentrifugation; circular dichroism (CD); dynamic light scattering (DLS); epidermal growth factor receptor (EGFR); hydrogen exchange mass spectrometry; intrinsically disordered protein; phosphotyrosine signaling; size exclusion chromatography; small angle X-ray scattering (SAXS).
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Franklin M. C., Carey K. D., Vajdos F. F., Leahy D. J., de Vos A. M., and Sliwkowski M. X. (2004) Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 5, 317–328 - PubMed
-
- Wood E. R., Truesdale A. T., McDonald O. B., Yuan D., Hassell A., Dickerson S. H., Ellis B., Pennisi C., Horne E., Lackey K., Alligood K. J., Rusnak D. W., Gilmer T. M., and Shewchuk L. (2004) A unique structure for epidermal growth factor receptor bound to GW572016 (lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 64, 6652–6659 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

