Structural and Biochemical Analyses Reveal the Mechanism of Glutathione S-Transferase Pi 1 Inhibition by the Anti-cancer Compound Piperlongumine
- PMID: 27872191
- PMCID: PMC5217671
- DOI: 10.1074/jbc.M116.750299
Structural and Biochemical Analyses Reveal the Mechanism of Glutathione S-Transferase Pi 1 Inhibition by the Anti-cancer Compound Piperlongumine
Abstract
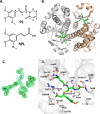
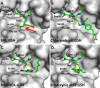
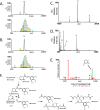
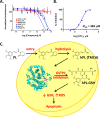
Glutathione S-transferase pi 1 (GSTP1) is frequently overexpressed in cancerous tumors and is a putative target of the plant compound piperlongumine (PL), which contains two reactive olefins and inhibits proliferation in cancer cells but not normal cells. PL exposure of cancer cells results in increased reactive oxygen species and decreased GSH. These data in tandem with other information led to the conclusion that PL inhibits GSTP1, which forms covalent bonds between GSH and various electrophilic compounds, through covalent adduct formation at the C7-C8 olefin of PL, whereas the C2-C3 olefin of PL was postulated to react with GSH. However, direct evidence for this mechanism has been lacking. To investigate, we solved the X-ray crystal structure of GSTP1 bound to PL and GSH at 1.1 Å resolution to rationalize previously reported structure activity relationship studies. Surprisingly, the structure showed that a hydrolysis product of PL (hPL) was conjugated to glutathione at the C7-C8 olefin, and this complex was bound to the active site of GSTP1; no covalent bond formation between hPL and GSTP1 was observed. Mass spectrometry (MS) analysis of the reactions between PL and GSTP1 confirmed that PL does not label GSTP1. Moreover, MS data also indicated that nucleophilic attack on PL at the C2-C3 olefin led to PL hydrolysis. Although hPL inhibits GSTP1 enzymatic activity in vitro, treatment of cells susceptible to PL with hPL did not have significant anti-proliferative effects, suggesting that hPL is not membrane-permeable. Altogether, our data suggest a model wherein PL is a prodrug whose intracellular hydrolysis initiates the formation of the hPL-GSH conjugate, which blocks the active site of and inhibits GSTP1 and thereby cancer cell proliferation.
Keywords: GSTP1; cancer; cancer therapy; crystal structure; inhibition; inhibition mechanism; natural product; piperlongumine; structure; therapeutic.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Raj L., Ide T., Gurkar A. U., Foley M., Schenone M., Li X., Tolliday N. J., Golub T. R., Carr S. A., Shamji A. F., Stern A. M., Mandinova A., Schreiber S. L., and Lee S. W. (2011) Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 475, 231–234 - PMC - PubMed
-
- Liu Y., Chang Y., Yang C., Sang Z., Yang T., Ang W., Ye W., Wei Y., Gong C., and Luo Y. (2014) Biodegradable nanoassemblies of piperlongumine display enhanced anti-angiogenesis and anti-tumor activities. Nanoscale 6, 4325–4337 - PubMed
-
- Bharadwaj U., Eckols T. K., Kolosov M., Kasembeli M. M., Adam A., Torres D., Zhang X., Dobrolecki L. E., Wei W., Lewis M. T., Dave B., Chang J. C., Landis M. D., Creighton C. J., Mancini M. A., and Tweardy D. J. (2015) Drug-repositioning screening identified piperlongumine as a direct STAT3 inhibitor with potent activity against breast cancer. Oncogene 34, 1341–1353 - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

